import warnings
from pathlib import Path
from urllib.parse import urlparse
import earthaccess
import xarray as xr
import virtualizarr as vz
from tqdm.notebook import tqdm
# VirtualiZarr components
from virtualizarr import open_virtual_dataset
from virtualizarr.parsers import HDFParser, KerchunkJSONParser
from virtualizarr.registry import ObjectStoreRegistry
# Object store components
from obstore.store import LocalStore, S3Store
from obstore.auth.earthdata import NasaEarthdataCredentialProviderPREFIRE Virtual Datacube Generation
Overview
Create Kerchunk reference files for cloud-based PREFIRE satellite data analysis without downloading files.
What You’ll Build:
- A 12MB Kerchunk file representing ~1GB+ of satellite data
- A virtual datacube that reads directly from cloud storage
- Visualizations from cloud data without full downloads
Prerequisites
- virtualizarr >= 2.0.1
- NASA Earthdata account (sign up here)
Virtual vs Traditional Approach
- Traditional: Download files (GB-TB) → Load into memory (or run out of memory) → Analyze
- Virtual: Create lightweight reference files (MB) → Access on-demand (without downloading) → Analyze at scale
1. Setup
Configure authentication and initialize essential libraries, including:
- earthaccess: NASA data search and authentication
- virtualizarr: Virtual dataset creation
- xarray: Multidimensional data handling
- obstore: Cloud storage access
from packaging import version
print(f"virtualizarr version: {vz.__version__}")
if version.parse(vz.__version__) < version.parse("2.0.1"):
print("⚠️ This notebook requires virtualizarr >= 2.0.1")
print(" Install with: pip install 'virtualizarr>=2.0.1'")virtualizarr version: 2.0.1try:
auth = earthaccess.login()
print(
f"✓ Authenticated as {auth.username}" if auth.authenticated else "❌ Authentication failed"
)
except Exception as e:
print(f"❌ Login failed: {e}")
print("Check credentials, or visit: https://urs.earthdata.nasa.gov/users/new for account setup")
raise✓ Authenticated as dkaufas# We can use the NASA credential provider to automatically refresh credentials
CREDENTIAL_URL = "https://data.asdc.earthdata.nasa.gov/s3credentials"
if not CREDENTIAL_URL.startswith("https://"):
raise ValueError("Credential URL must use HTTPS")
try:
cp = NasaEarthdataCredentialProvider(CREDENTIAL_URL)
print("✓ Credential provider initialized")
except Exception as e:
print(f"❌ Failed to initialize credential provider: {e}")
raise✓ Credential provider initialized2. Create Virtual Datacube
Create a virtual representation of PREFIRE data without downloading files:
- Search cloud-hosted NetCDF files
- Create virtual datasets for each file and group
- Combine into unified structure
PREFIRE Data Structure
- Root group (
""): Global metadata and coordinates - Sfc-Sorted group: Surface-sorted emission data
We’ll process both groups separately, then merge them.
2.0. Configuration
# Configuration - Modify these settings to customize the workflow
GROUP_NAMES = ["", "Sfc-Sorted"] # HDF5 groups to process
CONCATENATION_DIMENSION = "time" # Dimension to combine files along
OUTPUT_PATH = Path("prefire_kerchunk_20250819.json") # Output reference file
# Concatenation options
XARRAY_CONCAT_OPTIONS = {
"coords": "minimal", # Only include necessary coordinates
"compat": "override", # For non-concatenated variables of the same name, pick from first file
"combine_attrs": "override", # Use first file's attributes
# Note: "data_vars": "minimal" omitted because we're creating a new time dimension
}
print("🔧 Configuration:")
print(f" Processing {len(GROUP_NAMES)} groups: {GROUP_NAMES}")
print(f" Output file: {OUTPUT_PATH}")
print(f" Concatenation dimension: {CONCATENATION_DIMENSION}")🔧 Configuration:
Processing 2 groups: ['', 'Sfc-Sorted']
Output file: prefire_kerchunk_20250819.json
Concatenation dimension: time2.1. Search NASA Data Archive
Search for PREFIRE satellite files in NASA’s cloud storage.
# Retrieve data files for the dataset I'm interested in
print("Searching for PREFIRE Level 3 surface-sorted data...")
results = earthaccess.search_data(
short_name="PREFIRE_SAT2_3-SFC-SORTED-ALLSKY",
version="R01",
cloud_hosted=True,
# temporal=("2025-06-30 12:00", "2025-07-01 12:00"), # Uncomment to limit time range
)
if not results:
raise ValueError("No PREFIRE data found - check search parameters")
print(f"✓ Found {len(results)} granules")Searching for PREFIRE Level 3 surface-sorted data...
✓ Found 12 granules# Get S3 endpoints for all files:
s3_data_links = [g.data_links(access="direct")[0] for g in results]
parsed_s3_url = urlparse(s3_data_links[0])
print("First few granules: \n {}".format("\n ".join(s3_data_links[0:3])))
print(f"\nparsed = {parsed_s3_url}")
print(f"Bucket: {parsed_s3_url.netloc}")
s3_store = S3Store(
bucket=parsed_s3_url.netloc,
region="us-west-2",
credential_provider=cp,
virtual_hosted_style_request=False,
client_options={"allow_http": True},
)
object_registry = ObjectStoreRegistry({f"s3://{parsed_s3_url.netloc}": s3_store})
s3_storeFirst few granules:
s3://asdc-prod-protected/PREFIRE/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01/2024.07.01/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01_P00_20240701000000_20240731235959.nc
s3://asdc-prod-protected/PREFIRE/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01/2024.08.01/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01_P00_20240801000000_20240831235959.nc
s3://asdc-prod-protected/PREFIRE/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01/2024.09.01/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01_P00_20240901000000_20240930235959.nc
parsed = ParseResult(scheme='s3', netloc='asdc-prod-protected', path='/PREFIRE/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01/2024.07.01/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01_P00_20240701000000_20240731235959.nc', params='', query='', fragment='')
Bucket: asdc-prod-protectedS3Store(bucket="asdc-prod-protected")# # Un-comment this cell if you want to download the original files.
# files_downloaded = earthaccess.download(results[0])
# dtree = xr.open_datatree(files_downloaded[0])
# dtree2.2. Test a single file
def create_virtual_dataset(file_path: str, group_name: str):
"""Create virtual dataset from NetCDF group without loading data.
Without downloading files, build lightweight references to cloud data, containing:
- **Metadata**: Dimensions, coordinates, attributes
- **Chunk references**: Pointers to specific byte ranges in cloud storage
- **No actual data**: Just instructions on how to fetch it when needed
"""
with warnings.catch_warnings():
warnings.filterwarnings("ignore", message="Numcodecs codecs*", category=UserWarning)
return open_virtual_dataset(
file_path, parser=HDFParser(group=group_name), registry=object_registry
)results[0]Data: PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01_P00_20240701000000_20240731235959.nc
Size: 0 MB
Cloud Hosted: True
create_virtual_dataset(file_path=s3_data_links[0], group_name="Sfc-Sorted")<xarray.Dataset> Size: 16GB
Dimensions: (xtrack: 8, spectral: 63, sfc_type: 9, lat: 168,
lon: 360)
Dimensions without coordinates: xtrack, spectral, sfc_type, lat, lon
Data variables: (12/20)
wavelength (xtrack, spectral) float32 2kB ManifestArray<sh...
idealized_wavelength (xtrack, spectral) float32 2kB ManifestArray<sh...
surface_type_for_sorting (sfc_type) int8 9B ManifestArray<shape=(9,), dt...
latitude (lat, lon) float32 242kB ManifestArray<shape=(1...
longitude (lat, lon) float32 242kB ManifestArray<shape=(1...
emis_mean (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
... ...
emis_sum (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
asc_emis_sum (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
desc_emis_sum (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
emis_sumsquares (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
asc_emis_sumsquares (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
desc_emis_sumsquares (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...2.3. Process All Files
%%time
print(f"Processing {len(s3_data_links)} files across {len(GROUP_NAMES)} groups...")
vdatasets_by_group = {group: [] for group in GROUP_NAMES}
for file_index, file_path in enumerate(tqdm(s3_data_links, desc="Processing files")):
for group_name in tqdm(GROUP_NAMES, desc=f"Groups (file {file_index + 1})", leave=False):
virtual_ds = create_virtual_dataset(file_path, group_name)
vdatasets_by_group[group_name].append(virtual_ds)
print(f"✓ Created {sum(len(d) for d in vdatasets_by_group.values())} virtual datasets")Processing 12 files across 2 groups...✓ Created 24 virtual datasets
CPU times: user 9.5 s, sys: 15.4 s, total: 24.9 s
Wall time: 1min 57s# Show what a virtual dataset looks like
print("\nSample virtual dataset structure:")
sample_vds = vdatasets_by_group["Sfc-Sorted"][0]
sample_vds
Sample virtual dataset structure:<xarray.Dataset> Size: 16GB
Dimensions: (xtrack: 8, spectral: 63, sfc_type: 9, lat: 168,
lon: 360)
Dimensions without coordinates: xtrack, spectral, sfc_type, lat, lon
Data variables: (12/20)
wavelength (xtrack, spectral) float32 2kB ManifestArray<sh...
idealized_wavelength (xtrack, spectral) float32 2kB ManifestArray<sh...
surface_type_for_sorting (sfc_type) int8 9B ManifestArray<shape=(9,), dt...
latitude (lat, lon) float32 242kB ManifestArray<shape=(1...
longitude (lat, lon) float32 242kB ManifestArray<shape=(1...
emis_mean (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
... ...
emis_sum (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
asc_emis_sum (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
desc_emis_sum (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
emis_sumsquares (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
asc_emis_sumsquares (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...
desc_emis_sumsquares (xtrack, sfc_type, lat, lon, spectral) float32 1GB ManifestArray<shape=(8, 9, 168, 360, 63), dtype=float32, chunks=(2, 3, 56, 120, ...%%time
# Concatenate datasets for each group.
concatenated_by_group = {}
for group_name, group_datasets in vdatasets_by_group.items():
concatenated_by_group[group_name] = xr.concat(
group_datasets, dim=CONCATENATION_DIMENSION, **XARRAY_CONCAT_OPTIONS
)
print(f" ✓ Group '{group_name}': {concatenated_by_group[group_name].sizes}") ✓ Group '': Frozen({})/srv/conda/envs/notebook/lib/python3.11/site-packages/numcodecs/zarr3.py:133: UserWarning: Numcodecs codecs are not in the Zarr version 3 specification and may not be supported by other zarr implementations.
super().__init__(**codec_config) ✓ Group 'Sfc-Sorted': Frozen({'time': 12, 'xtrack': 8, 'spectral': 63, 'sfc_type': 9, 'lat': 168, 'lon': 360})
CPU times: user 341 ms, sys: 0 ns, total: 341 ms
Wall time: 339 ms%%time
# Merge the groups into one dataset.
consolidated_dataset = xr.merge([vds for _, vds in concatenated_by_group.items()])
print(f"✓ Consolidated dataset created with {len(consolidated_dataset.data_vars)} variables")
print(f" Time dimension: {consolidated_dataset.sizes.get('time', 'N/A')} steps")
consolidated_dataset✓ Consolidated dataset created with 20 variables
Time dimension: 12 steps
CPU times: user 859 μs, sys: 0 ns, total: 859 μs
Wall time: 846 μs<xarray.Dataset> Size: 198GB
Dimensions: (time: 12, xtrack: 8, spectral: 63, sfc_type: 9,
lat: 168, lon: 360)
Dimensions without coordinates: time, xtrack, spectral, sfc_type, lat, lon
Data variables: (12/20)
wavelength (time, xtrack, spectral) float32 24kB ManifestA...
idealized_wavelength (time, xtrack, spectral) float32 24kB ManifestA...
surface_type_for_sorting (time, sfc_type) int8 108B ManifestArray<shape=...
latitude (time, lat, lon) float32 3MB ManifestArray<shap...
longitude (time, lat, lon) float32 3MB ManifestArray<shap...
emis_mean (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB ManifestArray<shape=(12, 8, 9, 168, 360, 63), dtype=float32, chunks=(1, 2, 3, 56...
... ...
emis_sum (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB ManifestArray<shape=(12, 8, 9, 168, 360, 63), dtype=float32, chunks=(1, 2, 3, 56...
asc_emis_sum (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB ManifestArray<shape=(12, 8, 9, 168, 360, 63), dtype=float32, chunks=(1, 2, 3, 56...
desc_emis_sum (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB ManifestArray<shape=(12, 8, 9, 168, 360, 63), dtype=float32, chunks=(1, 2, 3, 56...
emis_sumsquares (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB ManifestArray<shape=(12, 8, 9, 168, 360, 63), dtype=float32, chunks=(1, 2, 3, 56...
asc_emis_sumsquares (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB ManifestArray<shape=(12, 8, 9, 168, 360, 63), dtype=float32, chunks=(1, 2, 3, 56...
desc_emis_sumsquares (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB ManifestArray<shape=(12, 8, 9, 168, 360, 63), dtype=float32, chunks=(1, 2, 3, 56...
Attributes: (12/26)
Conventions: CF-1.9
Format: netCDF-4
summary: The PREFIRE Level 3 products provide gridd...
full_versionID: R01_P00
archival_versionID: 01
granule_type: 3-SFC-SORTED-ALLSKY
... ...
UTC_coverage_end: 2024-07-31T23:59:59.969650
additional_file_description:
netCDF_lib_version: 4.9.2
orbit_sim_version:
SRF_NEdR_version:
is_this_a_final_product_file: yesFuture Development: build it into a DataTree
# # DataTree->Kerchunk is not yet implemented in virtualizarr
# # Will raise AttributeError: 'VirtualiZarrDataTreeAccessor' object has no attribute 'to_kerchunk'
# virtual_datatree = xr.DataTree.from_dict(each_group_concatenated)
# virtual_datatree2.4. Export Kerchunk
Future Development: also export to icechunk
print("Writing consolidated virtual dataset to Kerchunk format...")
try:
consolidated_dataset.vz.to_kerchunk(OUTPUT_PATH, format="json")
print(f"✓ Successfully wrote virtual dataset to {OUTPUT_PATH}")
print(f"File size: {OUTPUT_PATH.stat().st_size / (1024 * 1024):.1f} MB")
except Exception as e:
print(f"❌ Failed to write output file: {e}")
raiseWriting consolidated virtual dataset to Kerchunk format...
✓ Successfully wrote virtual dataset to prefire_kerchunk_20250819.json
File size: 11.6 MB3. Load Virtual Datacube
Read the Kerchunk file we created to demonstrate usage in a new session or for sharing with collaborators.
3.1. Setup
warnings.filterwarnings(
"ignore",
message="Numcodecs codecs are not in the Zarr version 3 specification*",
category=UserWarning,
)# Register the local file with our existing object registry
kerchunk_file_url = f"file://{Path.cwd() / OUTPUT_PATH.name}"
object_registry.register(kerchunk_file_url, LocalStore(prefix=Path.cwd()))
print(f"✓ Registered local Kerchunk file: {kerchunk_file_url}")✓ Registered local Kerchunk file: file:///home/jovyan/earthaccess-virtualizar/prefire_kerchunk_20250819.json3.2. Read reference file
json_parser = KerchunkJSONParser()
manifest_store = json_parser(url=kerchunk_file_url, registry=object_registry)
manifest_store<virtualizarr.manifests.store.ManifestStore at 0x7fa3c4411110>data = xr.open_zarr(manifest_store, consolidated=False, zarr_format=3)
data<xarray.Dataset> Size: 237GB
Dimensions: (time: 12, xtrack: 8, spectral: 63, sfc_type: 9,
lat: 168, lon: 360)
Dimensions without coordinates: time, xtrack, spectral, sfc_type, lat, lon
Data variables: (12/20)
wavelength (time, xtrack, spectral) float32 24kB dask.array<chunksize=(1, 8, 63), meta=np.ndarray>
idealized_wavelength (time, xtrack, spectral) float32 24kB dask.array<chunksize=(1, 8, 63), meta=np.ndarray>
surface_type_for_sorting (time, sfc_type) float32 432B dask.array<chunksize=(1, 9), meta=np.ndarray>
latitude (time, lat, lon) float32 3MB dask.array<chunksize=(1, 168, 360), meta=np.ndarray>
longitude (time, lat, lon) float32 3MB dask.array<chunksize=(1, 168, 360), meta=np.ndarray>
emis_mean (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB dask.array<chunksize=(1, 2, 3, 56, 120, 21), meta=np.ndarray>
... ...
emis_sum (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB dask.array<chunksize=(1, 2, 3, 56, 120, 21), meta=np.ndarray>
asc_emis_sum (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB dask.array<chunksize=(1, 2, 3, 56, 120, 21), meta=np.ndarray>
desc_emis_sum (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB dask.array<chunksize=(1, 2, 3, 56, 120, 21), meta=np.ndarray>
emis_sumsquares (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB dask.array<chunksize=(1, 2, 3, 56, 120, 21), meta=np.ndarray>
asc_emis_sumsquares (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB dask.array<chunksize=(1, 2, 3, 56, 120, 21), meta=np.ndarray>
desc_emis_sumsquares (time, xtrack, sfc_type, lat, lon, spectral) float32 13GB dask.array<chunksize=(1, 2, 3, 56, 120, 21), meta=np.ndarray>
Attributes: (12/26)
Conventions: CF-1.9
Format: netCDF-4
summary: The PREFIRE Level 3 products provide gridd...
full_versionID: R01_P00
archival_versionID: 01
granule_type: 3-SFC-SORTED-ALLSKY
... ...
UTC_coverage_end: 2024-07-31T23:59:59.969650
additional_file_description:
netCDF_lib_version: 4.9.2
orbit_sim_version:
SRF_NEdR_version:
is_this_a_final_product_file: yes4. Validate Results
Compare the virtual datacube output against an individual source file to verify data integrity.
from virtualizarr.utils import ObstoreReadersample_file_url = urlparse(s3_data_links[-1])
print(f"Comparing with individual file: {sample_file_url.path}")Comparing with individual file: /PREFIRE/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01/2025.06.01/PREFIRE_SAT2_3-SFC-SORTED-ALLSKY_R01_P00_20250601000000_20250630235959.nc# Create reader for direct file access
file_reader = ObstoreReader(s3_store, sample_file_url.path)# Load the same group directly for comparison
individual_dataset = xr.open_dataset(file_reader, engine="h5netcdf", group="Sfc-Sorted")# Compare dimensions between consolidated and individual datasets — we should have a new 'time' dimension
print("Validation Results:")
print(f" Individual file: {individual_dataset.sizes}")
print(f" Virtual datacube: {data.sizes}")
print(f" ✓ Successfully added time dimension: {data.sizes.get('time', 0)} timesteps")
symmetric_diff = dict(dict(individual_dataset.sizes).items() ^ dict(data.sizes).items())
print(f"Difference: {symmetric_diff}")
# Check that wavelengths match
if "wavelength" in individual_dataset.coords and "wavelength" in data.coords:
wavelengths_match = individual_dataset.wavelength.equals(data.wavelength)
print(f" ✓ Wavelength coordinates preserved: {wavelengths_match}")Validation Results:
Individual file: Frozen({'xtrack': 8, 'spectral': 63, 'sfc_type': 9, 'lat': 168, 'lon': 360})
Virtual datacube: Frozen({'time': 12, 'xtrack': 8, 'spectral': 63, 'sfc_type': 9, 'lat': 168, 'lon': 360})
✓ Successfully added time dimension: 12 timesteps
Difference: {'time': 12}5. Analyze and Visualize
Explore the virtual datacube structure and create a sample visualization of PREFIRE emissivity data.
5.1. Data Overview
print("📊 Dataset overview:")
if "summary" in data.attrs:
print(f" Description: {data.attrs['summary'][:100]}...")
print(f" Dimensions: {dict(data.sizes)}")
print(f" Data variables: {len(data.data_vars)}")
print("\n🔍 Key data variables:")
print(f" • emis_mean: shape:{data.emis_mean.shape} - Surface emission data")
print(f" • wavelength: {data.wavelength.min().values:.1f}-{data.wavelength.max().values:.1f} μm")
print(
f" • surface_type_for_sorting: {data.surface_type_for_sorting.flag_values} surface categories"
)
print(" • latitude/longitude: Geographic coordinates for each measurement")
print(f"\n⏱️ (new dimension) Time coverage: {data.sizes['time']} timesteps")📊 Dataset overview:
Description: The PREFIRE Level 3 products provide gridded, sorted, and/or statistical representations of other PR...
Dimensions: {'time': 12, 'xtrack': 8, 'spectral': 63, 'sfc_type': 9, 'lat': 168, 'lon': 360}
Data variables: 20
🔍 Key data variables:
• emis_mean: shape:(12, 8, 9, 168, 360, 63) - Surface emission data
• wavelength: 4.5-54.1 μm
• surface_type_for_sorting: [1, 2, 3, 4, 5, 6, 7, 8, 9] surface categories
• latitude/longitude: Geographic coordinates for each measurement
⏱️ (new dimension) Time coverage: 12 timesteps5.2. Create visualization
We will create a visualization to demonstrate virtual datacube functionality similar to xarray usage with a standard dataset.
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
from xarray.plot.utils import label_from_attrs# projection = ccrs.PlateCarree()
projection = ccrs.Orthographic(0, 90)
def make_nice_map(axis):
axis.add_feature(cfeature.OCEAN, color="lightblue")
axis.add_feature(cfeature.COASTLINE, edgecolor="grey")
axis.add_feature(cfeature.BORDERS, edgecolor="grey", linewidth=0.5)
grid = axis.gridlines(draw_labels=["left", "bottom"], dms=True, linestyle=":")
grid.xformatter = LONGITUDE_FORMATTER
grid.yformatter = LATITUDE_FORMATTER# Extract data for plotting
time_idx, xtrack_idx, surface_idx, spectral_idx = 0, 5, 7, 5
x = data["longitude"][time_idx, :, :].squeeze()
y = data["latitude"][time_idx, :, :].squeeze()
values = data.emis_mean[time_idx, xtrack_idx, surface_idx, :, :, spectral_idx].squeeze()fig, ax = plt.subplots(figsize=(10, 6), subplot_kw={"projection": projection})
make_nice_map(ax)
contour_handle = ax.contourf(
x,
y,
values,
transform=ccrs.PlateCarree(),
alpha=0.8,
zorder=2,
)
cb = plt.colorbar(contour_handle)
cb.set_label(label_from_attrs(values))
plt.show()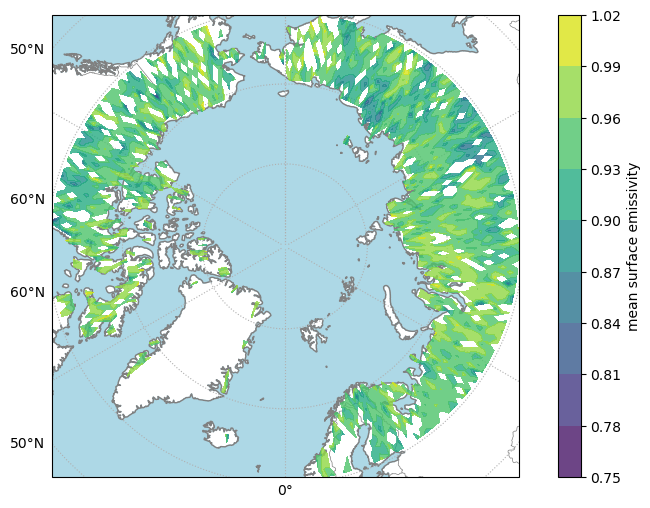
Summary
🎉 Congratulations! You’ve successfully created a cloud-native data analysis workflow.
What You Accomplished
- ✅ Built a virtual datacube from numerous satellite files without downloading
- ✅ Created a ~12MB reference file representing GB+ of data
- ✅ Analyzed and visualized satellite data directly from cloud storage
- ✅ Learned transferable skills for other cloud-hosted datasets
Next Steps
- Expand temporally: Add date ranges to process months/years of data
- Try other missions: Apply to MODIS, GOES, or other NASA datasets
- Add parallel processing: Use Dask for distributed computing
- Share your work: Export reference files for collaborators
Resources
📚 Kerchunk documentation • VirtualiZarr tutorials • NASA Earthdata Cloud

