import getpass
from datetime import (
datetime,
timezone,
) # needed to work with time in plotting time series
import earthaccess # needed to discover and download TEMPO data
import geopy.distance as geodist
import matplotlib.pyplot as plt # needed to plot the resulting time series
import netCDF4 as nc # needed to read TEMPO data
import numpy as np
from harmony import BBox, Client, Collection, Request
from harmony.config import EnvironmentPlotting timeseries from PREFIRE data
Summary
This notebook shows how to retrieve and plot time series of Polar Radiant Energy in the Far-InfraRed Experiment (PREFIRE) data for points of interest.
Prerequisites
- earthaccess
- geopy: the library is needed for determination of a true nearest neighbor observation
- matplotlib
- netCDF4
- numpy
- harmony-py
Importing the necessary libraries
Define a function to read FLX data
this function reads both original granules and subset
though the outputs have different dimensions due to different file structure of original and subset data files
def read_PREFIRE_2B_FLX(fn):
try:
with nc.Dataset(fn) as ds:
geo = ds.groups["Geometry"]
var = geo.variables["ctime"]
ctime = np.ma.getdata(var[:])
var = geo.variables["ctime_minus_UTC"]
ctime_minus_UTC = np.ma.getdata(var[:])
var = geo.variables["latitude"]
lat = np.ma.getdata(var[:])
var = geo.variables["longitude"]
lon = np.ma.getdata(var[:])
var = geo.variables["time_UTC_values"]
time_UTC = np.ma.getdata(var[:])
Flx = ds.groups["Flx"]
var = Flx.variables["flx_qc_bitflags"]
flxbitQF = np.ma.getdata(var[:])
var = Flx.variables["flx_quality_flag"]
flxQF = np.ma.getdata(var[:])
var = Flx.variables["olr"]
olr = np.ma.getdata(var[:])
fv_olr = var.get_fill_value()
olr_unit = var.getncattr("units")
except Exception:
ctime = 0.0
ctime_minus_UTC = 0.0
lat = 0.0
lon = 0.0
time_UTC = 0.0
flxbitQF = 0.0
flxQF = 0.0
olr = 0.0
fv_olr = 0.0
olr_unit = 0.0
return ctime, ctime_minus_UTC, lat, lon, time_UTC, flxbitQF, flxQF, olr, fv_olr, olr_unitSet name constants
out_Q = "OLR"
out_Q_unit = "W m-2"Logging in with Earthdata credentials
# User needs to create an account at https://www.earthdata.nasa.gov/
# Function earthaccess.login prompts for EarthData login and password.
auth = earthaccess.login(strategy="interactive", persist=True)Define a function to obtain collection ID
collection ID will be needed for accessing the collection with Harmony-py
def get_collectionID_earthaccess(short_name, version):
collectionID = "-1"
results = earthaccess.search_datasets(short_name=short_name, version=version)
if len(results) == 1:
collectionID = results[0]["meta"]["concept-id"]
else:
raise Exception("Specify valid collection")
return collectionIDshort_name = "PREFIRE_SAT2_2B-FLX"
version = "R01"
collectionID = get_collectionID_earthaccess(short_name, version)
print(
f"collectionID for collection short_name {short_name} and version {version} is {collectionID}"
)collectionID for collection short_name PREFIRE_SAT2_2B-FLX and version R01 is C3499202317-LARC_CLOUDSearch for all granules covering point of interest (POI)
Set POI
Edge Island, Svalbard, Norway
POI_name = "Svalbard"
[POI_lon, POI_lat] = [22.5, 77.75]
POI = [POI_lon, POI_lat]
# POI location string for output files
if POI_lon < 0.0:
lon_str = str("%08.4fW" % (-POI_lon))
else:
lon_str = str("%08.4fE" % (POI_lon))
if POI_lat < 0.0:
lat_str = str("%08.4fS" % (-POI_lat))
else:
lat_str = str("%08.4fN" % (POI_lat))
POI_loc_str = POI_name + "_" + lat_str + "_" + lon_str
print(POI_loc_str)Svalbard_077.7500N_022.5000ESearch for granules through entire mission lifetime
short_name = "PREFIRE_SAT2_2B-FLX" # collection name
version = "R01" # version of the collection
results_earthaccess = earthaccess.search_data(
short_name=short_name, version=version, point=(POI_lon, POI_lat)
) # longitude-latitude pair
PREFIRE_names = sorted([r["meta"]["native-id"] for r in results_earthaccess])
print(len(PREFIRE_names), "granules were found")344 granules were foundLogging in with Harmony-py
print("Please provide your Earthdata Login credentials to allow data access")
print("Your credentials will only be passed to Earthdata and will not be exposed in the notebook")
username = input("Username:")
harmony_client = Client(env=Environment.PROD, auth=(username, getpass.getpass()))Please provide your Earthdata Login credentials to allow data access
Your credentials will only be passed to Earthdata and will not be exposed in the notebookUsername: alexrad71
········Define function creating Harmony-py request and returning job ID
def create_Harmony_job_id(filenames, collectionID, POI_lon, POI_lat):
# function create_Harmony_job_id takes a list of granule names to subset, filenames,
# a collection ID,
# and a position of the POI.
# the list of variables to retain is pre-defined
request = Request(
collection=Collection(id=collectionID),
granule_name=filenames,
variables=[
"Geometry/ctime",
"Geometry/ctime_minus_UTC",
"Geometry/latitude",
"Geometry/longitude",
"Geometry/time_UTC_values",
"Flx/flx_qc_bitflags",
"Flx/flx_quality_flag",
"Flx/olr",
],
# subsetting in space around the POI
spatial=BBox(POI_lon - 0.5, POI_lat - 0.5, POI_lon + 0.5, POI_lat + 0.5),
)
assert request.is_valid()
job_id = harmony_client.submit(request)
return job_idSearch and download subset data
un-documented “feature” of Request function used in the function definition above is that
the length of the list granules is limited. It is somewhere between 75 and 100.
We have to split a longer list into parts.
chunk_size = 75
num_files = len(PREFIRE_names)
print(num_files)
all_filenames = [] * num_files
all_filenames[:] = PREFIRE_names[:]
job_id_list = []
i = 0
while len(all_filenames) > chunk_size:
chunk = all_filenames[:chunk_size]
# print(chunk)
del all_filenames[:chunk_size]
job_id = create_Harmony_job_id(chunk, collectionID, POI_lon, POI_lat)
job_id_list.append(job_id)
i = i + 1
print("chunk ", i, " job_id:", job_id)
last_chunk = all_filenames
last_job_id = create_Harmony_job_id(last_chunk, collectionID, POI_lon, POI_lat)
print("chunk ", i + 1, " job_id:", last_job_id)
job_id_list.append(last_job_id)
print(len(PREFIRE_names))
print(len(all_filenames))
all_results_stored = []
for job_id in job_id_list:
print(f"jobID = {job_id}")
harmony_client.wait_for_processing(job_id, show_progress=True)
# Download the resulting files
results = harmony_client.download_all(job_id, directory="./", overwrite=True)
all_results_stored.append([f.result() for f in results])
print(len(all_results_stored))
fout = open("subset_names.txt", "w")
for result in all_results_stored:
for res in result:
fout.write(res + "\n")
fout.close()344
chunk 1 job_id: 576a333e-11d5-4b04-83d3-28290231078e
chunk 2 job_id: 8d657e5d-4093-436e-83a6-babf0edaf33f
chunk 3 job_id: 1cfb7896-aaa0-4a27-90dd-500a13198959
chunk 4 job_id: 49dcb55d-263b-444f-bbc4-6e840e06fb45 [ Processing: 0% ] | | [/]chunk 5 job_id: 887d50ba-2c5a-4b76-99ad-f9925009f1f4
344
44
jobID = 576a333e-11d5-4b04-83d3-28290231078e [ Processing: 100% ] |###################################################| [|]./101597224_PREFIRE_SAT2_2B-FLX_R01_P00_20240630033949_00541_subsetted_20240831T032430Z_C3499202317-LARC_CLOUD_merged.nc4
jobID = 8d657e5d-4093-436e-83a6-babf0edaf33f [ Processing: 100% ] |###################################################| [|]
[ Processing: 0% ] | | [/]./101597223_PREFIRE_SAT2_2B-FLX_R01_P00_20240831205207_01489_subsetted_20241109T210231Z_C3499202317-LARC_CLOUD_merged.nc4
jobID = 1cfb7896-aaa0-4a27-90dd-500a13198959 [ Processing: 100% ] |###################################################| [|]
[ Processing: 0% ] | | [/]./101597221_PREFIRE_SAT2_2B-FLX_R01_P00_20241110032306_02552_subsetted_20250114T031335Z_C3499202317-LARC_CLOUD_merged.nc4
jobID = 49dcb55d-263b-444f-bbc4-6e840e06fb45 [ Processing: 100% ] |###################################################| [|]
[ Processing: 0% ] | | [/]./101597222_PREFIRE_SAT2_2B-FLX_R01_P00_20250114203927_03547_subsetted_20250327T032736Z_C3499202317-LARC_CLOUD_merged.nc4
jobID = 887d50ba-2c5a-4b76-99ad-f9925009f1f4 [ Processing: 100% ] |###################################################| [|]./101597219_PREFIRE_SAT2_2B-FLX_R01_P00_20250327205240_04638_subsetted_20250509T033927Z_C3499202317-LARC_CLOUD_merged.nc4
5
./101597654_PREFIRE_SAT2_2B-FLX_R01_P00_20240630002914_00539_subsetted_20240915T003053Z_C3499202317-LARC_CLOUD_merged.nc4
./101597663_PREFIRE_SAT2_2B-FLX_R01_P00_20240915082659_01708_subsetted_20241214T083928Z_C3499202317-LARC_CLOUD_merged.nc4
./101597653_PREFIRE_SAT2_2B-FLX_R01_P00_20241215003034_03080_subsetted_20250318T085053Z_C3499202317-LARC_CLOUD_merged.nc4
./101597652_PREFIRE_SAT2_2B-FLX_R01_P00_20250319004104_04504_subsetted_20250509T002931Z_C3499202317-LARC_CLOUD_merged.nc4Create timeseries of OLR
clear and cloudy retrievals are separated by quality flag
nearest neighbor are limited to be less than 25 km avay from the POI, see figures of atrack and xtrack distances distributions below.
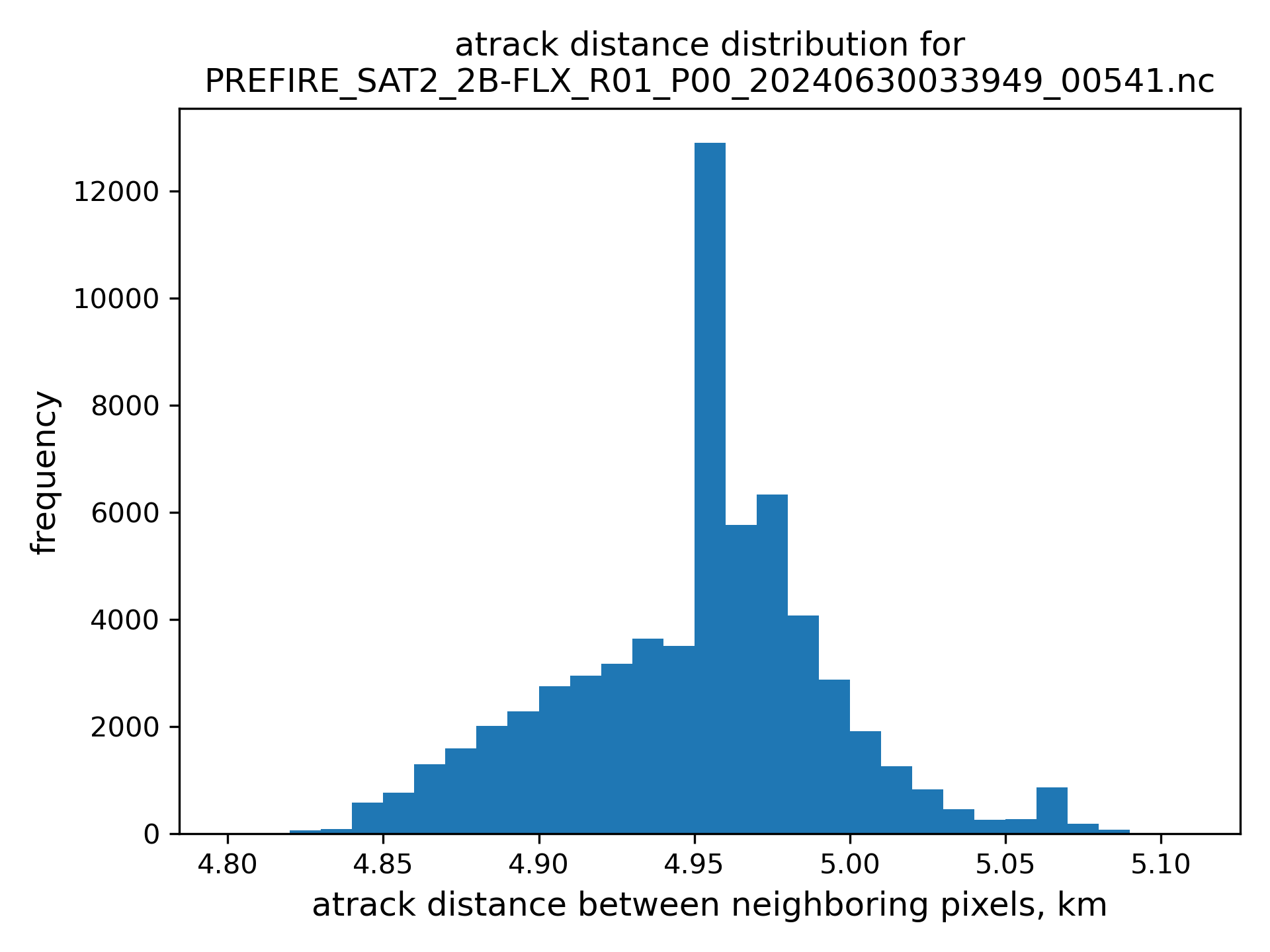
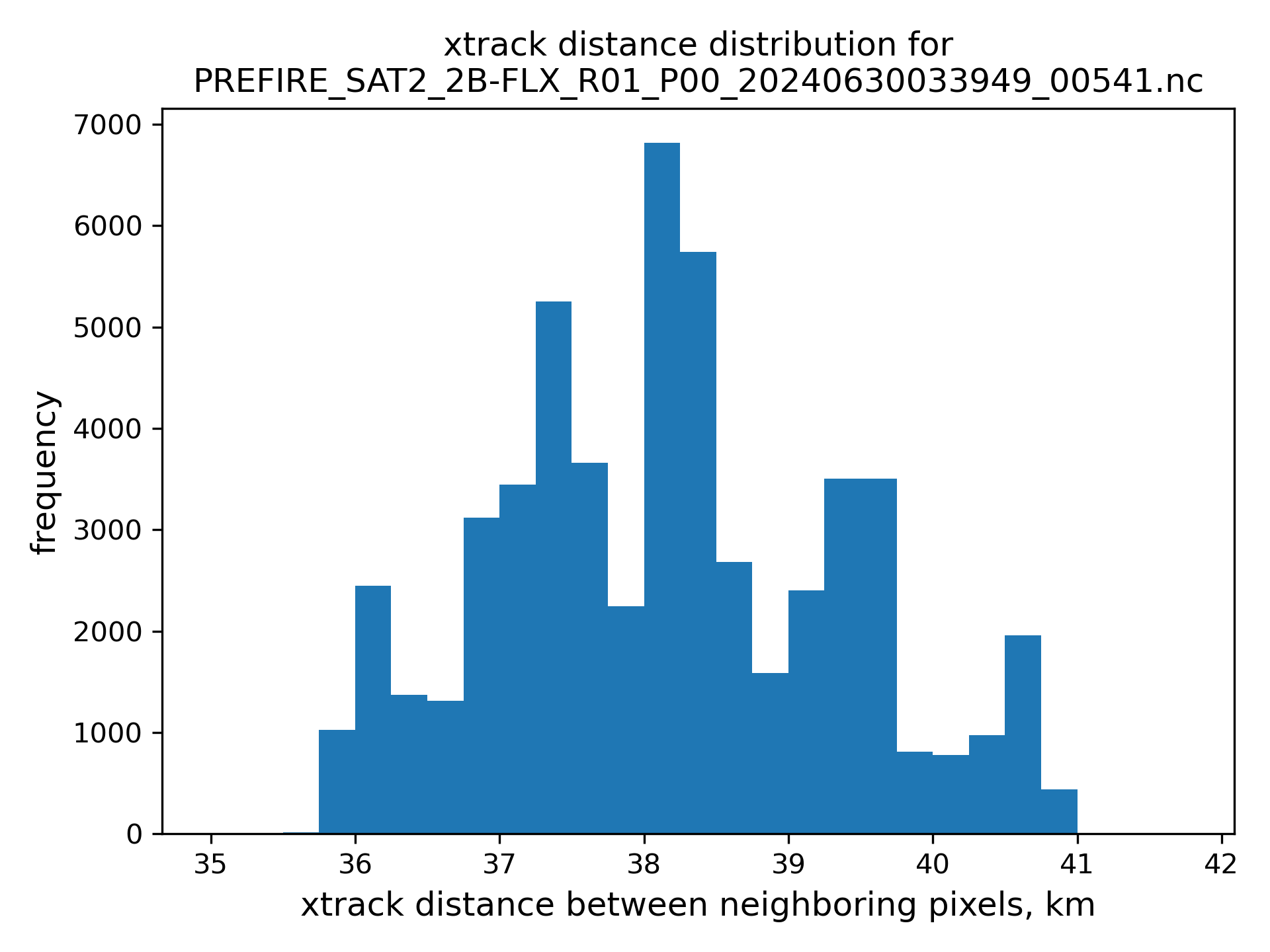
# read subset file names back
fin = open("subset_names.txt", "r")
subset_filenames = fin.readlines()
fin.close()
fout_HQ_clear = open(out_Q + "_clear_" + POI_loc_str, "w")
fout_HQ_clear.write("timeseries of clear" + out_Q + " at " + POI_loc_str + "\n")
fout_HQ_clear.write("YYYY MM DD hh mm ss ms lat lon olr," + out_Q_unit + " dist_to_POI\n")
fout_HQ_cloudy = open(out_Q + "_cloudy_" + POI_loc_str, "w")
fout_HQ_cloudy.write("timeseries of cloudy" + out_Q + " at " + POI_loc_str + "\n")
fout_HQ_cloudy.write(
"YYYY MM DD hh mm ss ms lat lon olr," + out_Q_unit + " dist_to_POI\n"
)
for fn in subset_filenames:
# the subsets are in local directory, so we need only file name, .split('/')[-1] removes path, [:-1] remove "new line" at the end
fname = fn.split("/")[-1][:-1]
print(fname)
ctime, ctime_minus_UTC, lat, lon, time_UTC, flxbitQF, flxQF, olr, fv_olr, olr_unit = (
read_PREFIRE_2B_FLX(fname)
)
# subset files consist of multiple granule subsets, check the number of granules contributing to the subset file
ngranules = ctime.shape[0]
for igranule in range(ngranules):
mask_hq = (olr[igranule] != fv_olr) & (flxbitQF[igranule] == 0)
# search for clear-sky observations
mask_clear = mask_hq & (flxQF[igranule] == 0)
lat_clear = lat[igranule, mask_clear]
lon_clear = lon[igranule, mask_clear]
olr_clear = olr[igranule, mask_clear]
nclear = len(lat_clear)
if nclear > 0:
dist_clear = np.empty(nclear)
for i, (lat_loc, lon_loc) in enumerate(zip(lat_clear, lon_clear)):
dist_clear[i] = geodist.geodesic((POI_lat, POI_lon), (lat_loc, lon_loc)).km
olr_clear_to_sort = np.stack((lat_clear, lon_clear, olr_clear, dist_clear), axis=1)
olr_clear_sorted = olr_clear_to_sort[olr_clear_to_sort[:, 3].argsort()]
if (
olr_clear_sorted[0, 3] <= 25.0
): # check whether the nearest pixel is within 25 km from the POI
atrack_index_clear = np.argwhere(lat[igranule] == olr_clear_sorted[0, 0])[0, 0]
time_clear = time_UTC[igranule, atrack_index_clear]
fout_HQ_clear.write(
f"{time_clear[0]:4d} {time_clear[1]:2d} {time_clear[2]:2d} {time_clear[3]:2d} {time_clear[4]:2d} {time_clear[5]:2d} {time_clear[6]:3d}"
)
fout_HQ_clear.write(
f"{olr_clear_sorted[0, 0]:8.4f} {olr_clear_sorted[0, 1]:8.4f} {olr_clear_sorted[0, 2]:8.4f} {olr_clear_sorted[0, 3]:8.4f}\n"
)
# search for cloudy observations
mask_cloudy = mask_hq & (flxQF[igranule] == 1)
lat_cloudy = lat[igranule, mask_cloudy]
lon_cloudy = lon[igranule, mask_cloudy]
olr_cloudy = olr[igranule, mask_cloudy]
ncloudy = len(lat_cloudy)
if ncloudy > 0:
dist_cloudy = np.empty(ncloudy)
for i, (lat_loc, lon_loc) in enumerate(zip(lat_cloudy, lon_cloudy)):
dist_cloudy[i] = geodist.geodesic((POI_lat, POI_lon), (lat_loc, lon_loc)).km
olr_cloudy_to_sort = np.stack((lat_cloudy, lon_cloudy, olr_cloudy, dist_cloudy), axis=1)
olr_cloudy_sorted = olr_cloudy_to_sort[olr_cloudy_to_sort[:, 3].argsort()]
if (
olr_cloudy_sorted[0, 3] <= 25.0
): # check whether the nearest pixel is within 25 km from the POI
atrack_index_cloudy = np.argwhere(lat[igranule] == olr_cloudy_sorted[0, 0])[0, 0]
time_cloudy = time_UTC[igranule, atrack_index_cloudy]
fout_HQ_cloudy.write(
f"{time_cloudy[0]:4d} {time_cloudy[1]:2d} {time_cloudy[2]:2d} {time_cloudy[3]:2d} {time_cloudy[4]:2d} {time_cloudy[5]:2d} {time_cloudy[6]:3d}"
)
fout_HQ_cloudy.write(
f"{olr_cloudy_sorted[0, 0]:8.4f} {olr_cloudy_sorted[0, 1]:8.4f} {olr_cloudy_sorted[0, 2]:8.4f} {olr_cloudy_sorted[0, 3]:8.4f}\n"
)
fout_HQ_clear.close()
fout_HQ_cloudy.close()101597224_PREFIRE_SAT2_2B-FLX_R01_P00_20240630033949_00541_subsetted_20240831T032430Z_C3499202317-LARC_CLOUD_merged.nc4
101597223_PREFIRE_SAT2_2B-FLX_R01_P00_20240831205207_01489_subsetted_20241109T210231Z_C3499202317-LARC_CLOUD_merged.nc4
101597221_PREFIRE_SAT2_2B-FLX_R01_P00_20241110032306_02552_subsetted_20250114T031335Z_C3499202317-LARC_CLOUD_merged.nc4
101597222_PREFIRE_SAT2_2B-FLX_R01_P00_20250114203927_03547_subsetted_20250327T032736Z_C3499202317-LARC_CLOUD_merged.nc4
101597219_PREFIRE_SAT2_2B-FLX_R01_P00_20250327205240_04638_subsetted_20250509T033927Z_C3499202317-LARC_CLOUD_merged.nc4define a function to read written down timeseries
def read_timeseries(ts_fname, yyyy_ini, mm_ini, dd_ini):
fout = open(ts_fname, "r")
_ = fout.readline() # header 1
_ = fout.readline() # header 2
data_lines = fout.readlines()
fout.close()
hh = 0
mm = 0
ss = 0
dt0 = datetime(yyyy_ini, mm_ini, dd_ini, hh, mm, ss, tzinfo=timezone.utc)
time_series = np.empty([0, 2])
for line in data_lines:
split = line.split()
YYYY = int(split[0])
MM = int(split[1])
DD = int(split[2])
hh = int(split[3])
mm = int(split[4])
ss = int(split[5])
ms = int(split[6])
if ms > 999:
ms = 999
olr = float(split[9])
obs_time = datetime(YYYY, MM, DD, hh, mm, ss, ms * 1000, tzinfo=timezone.utc)
# dt below is time since the beginning of the period of interest in days
dt = (obs_time - dt0).total_seconds() / 86400.0
time_series = np.append(time_series, [[dt, olr]], axis=0)
return time_series, dt0Read timeseries back
fname_clear = out_Q + "_clear_" + POI_loc_str
fname_cloudy = out_Q + "_cloudy_" + POI_loc_str
yyyy_ini = 2024
mm_ini = 6
dd_ini = 30
time_series_clear, dt0 = read_timeseries(fname_clear, yyyy_ini, mm_ini, dd_ini)
time_series_cloudy, dt0 = read_timeseries(fname_cloudy, yyyy_ini, mm_ini, dd_ini)Plot timeseries
plot_title = out_Q + " " + POI_loc_str
img_name = out_Q + "_" + POI_loc_str + ".jpg"
plt.plot(time_series_clear[:, 0], time_series_clear[:, 1], label="clear", c="r")
plt.plot(time_series_cloudy[:, 0], time_series_cloudy[:, 1], label="cloudy", c="b")
# Set the range of x-axis
l_lim = 0.0
u_lim = np.ceil(max(time_series_clear[-1, 0], time_series_cloudy[-1, 0]))
plt.xlim(l_lim, u_lim)
# some research is required to set the vertical range
plt.xlabel(r"GMT, day from " + dt0.strftime("%Y-%m-%d %H:%M:%S"), fontsize=12)
plt.ylabel("OLR, " + olr_unit, fontsize=12)
plt.legend(loc="lower left")
plt.title(plot_title)
plt.savefig(img_name, format="jpg", dpi=300)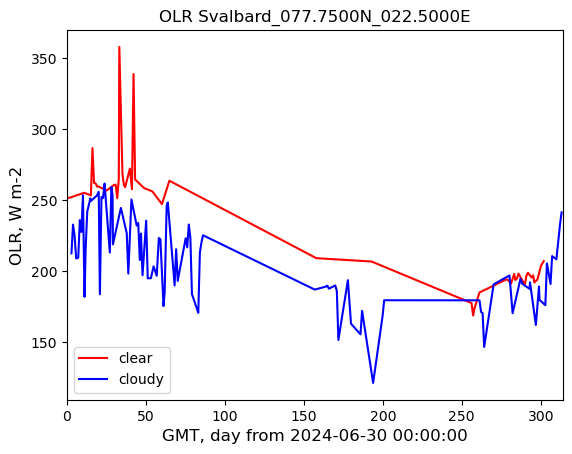
Set another POI
Dome C, Antarctica
# POI: Dome C, Antarctica
# Coordinates: 75°05′59″S 123°19′56″E
POI_lat = -(75 + 5 / 60.0 + 59 / 3600.0)
POI_lon = 123 + 19 / 60.0 + 56 / 3600.0
POI_name = "Dome_C"
# POI location string for output files
if POI_lon < 0.0:
lon_str = str("%08.4fW" % (-POI_lon))
else:
lon_str = str("%08.4fE" % (POI_lon))
if POI_lat < 0.0:
lat_str = str("%08.4fS" % (-POI_lat))
else:
lat_str = str("%08.4fN" % (POI_lat))
POI_loc_str = POI_name + "_" + lat_str + "_" + lon_str
print(POI_loc_str)Dome_C_075.0997S_123.3322ESearch for granules through entire mission lifetime
short_name = "PREFIRE_SAT2_2B-FLX" # collection name
version = "R01" # version of the collection
results_earthaccess = earthaccess.search_data(
short_name=short_name, version=version, point=(POI_lon, POI_lat)
) # longitude-latitude pair
PREFIRE_names = sorted([r["meta"]["native-id"] for r in results_earthaccess])print(len(PREFIRE_names))262Search and download subset data
un-documented “feature” of Request function used in the definition of function create_Harmony_job_id is that
the length of the list granules is limited. It is somewhere between 75 and 100.
We have to split a longer list into parts.
chunk_size = 75
num_files = len(PREFIRE_names)
print(num_files)
all_filenames = [] * num_files
all_filenames[:] = PREFIRE_names[:]
job_id_list = []
i = 0
while len(all_filenames) > chunk_size:
chunk = all_filenames[:chunk_size]
# print(chunk)
del all_filenames[:chunk_size]
job_id = create_Harmony_job_id(chunk, collectionID, POI_lon, POI_lat)
job_id_list.append(job_id)
i = i + 1
print("chunk ", i, " job_id:", job_id)
last_chunk = all_filenames
last_job_id = create_Harmony_job_id(last_chunk, collectionID, POI_lon, POI_lat)
print("chunk ", i + 1, " job_id:", last_job_id)
job_id_list.append(last_job_id)
print(len(PREFIRE_names))
print(len(all_filenames))
all_results_stored = []
for job_id in job_id_list:
print(f"jobID = {job_id}")
harmony_client.wait_for_processing(job_id, show_progress=True)
# Download the resulting files
results = harmony_client.download_all(job_id, directory="./", overwrite=True)
all_results_stored.append([f.result() for f in results])
print(len(all_results_stored))
fout = open("subset_names_Dome_C.txt", "w")
for result in all_results_stored:
for res in result:
fout.write(res + "\n")
fout.close()262
chunk 1 job_id: 13574ab4-7da6-4214-a924-43cb4fdde64c
chunk 2 job_id: e6487055-e043-499c-910e-fdad24d12f0c
chunk 3 job_id: b3ed9cd6-076f-48ca-b7c2-6690fdf45503 [ Processing: 0% ] | | [/]chunk 4 job_id: c20763ce-d0c7-4fbb-bc92-6af61dcf448b
262
37
jobID = 13574ab4-7da6-4214-a924-43cb4fdde64c [ Processing: 100% ] |###################################################| [|]jobID = e6487055-e043-499c-910e-fdad24d12f0c [ Processing: 100% ] |###################################################| [|]
[ Processing: 0% ] | | [/]jobID = b3ed9cd6-076f-48ca-b7c2-6690fdf45503 [ Processing: 100% ] |###################################################| [|]
[ Processing: 0% ] | | [/]jobID = c20763ce-d0c7-4fbb-bc92-6af61dcf448b [ Processing: 100% ] |###################################################| [|]4Create timeseries of OLR
clear and cloudy retrievals are separated by quality flag
nearest neighbor are limited to be less than 25 km avay from the POI.
# read subset file names back
fin = open("subset_names_Dome_C.txt", "r")
subset_filenames = fin.readlines()
fin.close()
fout_HQ_clear = open(out_Q + "_clear_" + POI_loc_str, "w")
fout_HQ_clear.write("timeseries of clear" + out_Q + " at " + POI_loc_str + "\n")
fout_HQ_clear.write("YYYY MM DD hh mm ss ms lat lon olr," + out_Q_unit + " dist_to_POI\n")
fout_HQ_cloudy = open(out_Q + "_cloudy_" + POI_loc_str, "w")
fout_HQ_cloudy.write("timeseries of cloudy" + out_Q + " at " + POI_loc_str + "\n")
fout_HQ_cloudy.write(
"YYYY MM DD hh mm ss ms lat lon olr," + out_Q_unit + " dist_to_POI\n"
)
for fn in subset_filenames:
# the subsets are in local directory, so we need only file name, .split('/')[-1] removes path, [:-1] remove "new line" at the end
fname = fn.split("/")[-1][:-1]
print(fname)
ctime, ctime_minus_UTC, lat, lon, time_UTC, flxbitQF, flxQF, olr, fv_olr, olr_unit = (
read_PREFIRE_2B_FLX(fname)
)
# subset files consist of multiple granule subsets, check the number of granules contributing to the subset file
ngranules = ctime.shape[0]
for igranule in range(ngranules):
mask_hq = (olr[igranule] != fv_olr) & (flxbitQF[igranule] == 0)
# search for clear-sky observations
mask_clear = mask_hq & (flxQF[igranule] == 0)
lat_clear = lat[igranule, mask_clear]
lon_clear = lon[igranule, mask_clear]
olr_clear = olr[igranule, mask_clear]
nclear = len(lat_clear)
if nclear > 0:
dist_clear = np.empty(nclear)
for i, (lat_loc, lon_loc) in enumerate(zip(lat_clear, lon_clear)):
dist_clear[i] = geodist.geodesic((POI_lat, POI_lon), (lat_loc, lon_loc)).km
olr_clear_to_sort = np.stack((lat_clear, lon_clear, olr_clear, dist_clear), axis=1)
olr_clear_sorted = olr_clear_to_sort[olr_clear_to_sort[:, 3].argsort()]
if (
olr_clear_sorted[0, 3] <= 25.0
): # check whether the nearest pixel is within 25 km from the POI
atrack_index_clear = np.argwhere(lat[igranule] == olr_clear_sorted[0, 0])[0, 0]
time_clear = time_UTC[igranule, atrack_index_clear]
fout_HQ_clear.write(
f"{time_clear[0]:4d} {time_clear[1]:2d} {time_clear[2]:2d} {time_clear[3]:2d} {time_clear[4]:2d} {time_clear[5]:2d} {time_clear[6]:3d}"
)
fout_HQ_clear.write(
f" {olr_clear_sorted[0, 0]:8.4f} {olr_clear_sorted[0, 1]:8.4f} {olr_clear_sorted[0, 2]:8.4f} {olr_clear_sorted[0, 3]:8.4f}\n"
)
# search for cloudy observations
mask_cloudy = mask_hq & (flxQF[igranule] == 1)
lat_cloudy = lat[igranule, mask_cloudy]
lon_cloudy = lon[igranule, mask_cloudy]
olr_cloudy = olr[igranule, mask_cloudy]
ncloudy = len(lat_cloudy)
if ncloudy > 0:
dist_cloudy = np.empty(ncloudy)
for i, (lat_loc, lon_loc) in enumerate(zip(lat_cloudy, lon_cloudy)):
dist_cloudy[i] = geodist.geodesic((POI_lat, POI_lon), (lat_loc, lon_loc)).km
olr_cloudy_to_sort = np.stack((lat_cloudy, lon_cloudy, olr_cloudy, dist_cloudy), axis=1)
olr_cloudy_sorted = olr_cloudy_to_sort[olr_cloudy_to_sort[:, 3].argsort()]
if (
olr_cloudy_sorted[0, 3] <= 25.0
): # check whether the nearest pixel is within 25 km from the POI
atrack_index_cloudy = np.argwhere(lat[igranule] == olr_cloudy_sorted[0, 0])[0, 0]
time_cloudy = time_UTC[igranule, atrack_index_cloudy]
fout_HQ_cloudy.write(
f"{time_cloudy[0]:4d} {time_cloudy[1]:2d} {time_cloudy[2]:2d} {time_cloudy[3]:2d} {time_cloudy[4]:2d} {time_cloudy[5]:2d} {time_cloudy[6]:3d}"
)
fout_HQ_cloudy.write(
f" {olr_cloudy_sorted[0, 0]:8.4f} {olr_cloudy_sorted[0, 1]:8.4f} {olr_cloudy_sorted[0, 2]:8.4f} {olr_cloudy_sorted[0, 3]:8.4f}\n"
)
fout_HQ_clear.close()
fout_HQ_cloudy.close()101597654_PREFIRE_SAT2_2B-FLX_R01_P00_20240630002914_00539_subsetted_20240915T003053Z_C3499202317-LARC_CLOUD_merged.nc4
101597663_PREFIRE_SAT2_2B-FLX_R01_P00_20240915082659_01708_subsetted_20241214T083928Z_C3499202317-LARC_CLOUD_merged.nc4
101597653_PREFIRE_SAT2_2B-FLX_R01_P00_20241215003034_03080_subsetted_20250318T085053Z_C3499202317-LARC_CLOUD_merged.nc4
101597652_PREFIRE_SAT2_2B-FLX_R01_P00_20250319004104_04504_subsetted_20250509T002931Z_C3499202317-LARC_CLOUD_merged.nc4Read timeseries back
fname_clear = out_Q + "_clear_" + POI_loc_str
fname_cloudy = out_Q + "_cloudy_" + POI_loc_str
yyyy_ini = 2024
mm_ini = 6
dd_ini = 30
time_series_clear, dt0 = read_timeseries(fname_clear, yyyy_ini, mm_ini, dd_ini)
time_series_cloudy, dt0 = read_timeseries(fname_cloudy, yyyy_ini, mm_ini, dd_ini)Plot timeseries
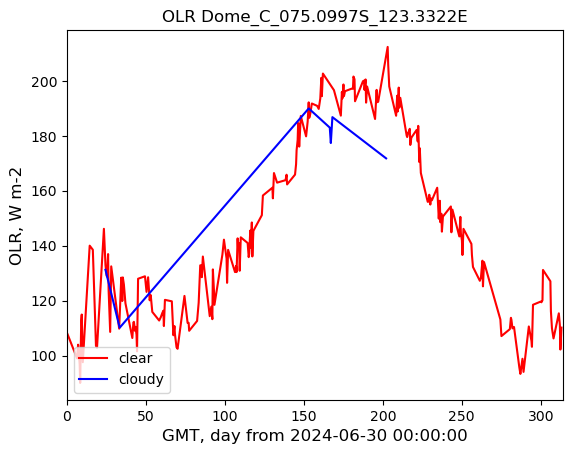
plot_title = out_Q + " " + POI_loc_str
img_name = out_Q + "_" + POI_loc_str + ".jpg"
plt.plot(time_series_clear[:, 0], time_series_clear[:, 1], label="clear", c="r")
plt.plot(time_series_cloudy[:, 0], time_series_cloudy[:, 1], label="cloudy", c="b")
# Set the range of x-axis
l_lim = 0.0
u_lim = np.ceil(max(time_series_clear[-1, 0], time_series_cloudy[-1, 0]))
plt.xlim(l_lim, u_lim)
# some research is required to set the vertical range
plt.xlabel(r"GMT, day from " + dt0.strftime("%Y-%m-%d %H:%M:%S"), fontsize=12)
plt.ylabel("OLR, " + olr_unit, fontsize=12)
plt.legend(loc="lower left")
plt.title(plot_title)
plt.savefig(img_name, format="jpg", dpi=300)