Exploring nitrogen dioxide (NO2) data from PACE/OCI#
Author(s): Anna Windle (NASA, SSAI), Zachary Fasnacht (NASA, SSAI)
Last updated: March 5, 2026
The following notebooks are prerequisites for this tutorial.
An Earthdata Login account is required to access data from the NASA Earthdata system, including NASA ocean color data.
Summary#
This tutorial describes how to access and download trace gas data products developed from PACE OCI data. More information on how these products were created can be found in Fasnacht et al. (2025). This notebook includes examples on how to plot L3M NO2 data as a static and interactive map, as well as how to plot an interactive time series plot. It also provides examples on how to open and plot L2 NO2 data.
Learning Objectives#
At the end of this notebook you will know:
How to access PACE OCI trace gas data products
Where to find high NO2 vertical column retrievals (hint: it’s a big city)
How to create a time series of NO2 data collected at a single location
Open and plot L2 NO2 data
1. Setup#
Begin by importing all of the packages used in this notebook and setting your Earthdata Login credentials.
from datetime import datetime
import cartopy
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import earthaccess
import holoviews
import hvplot.xarray
import matplotlib.colors
import matplotlib.pyplot as plt
import pandas as pd
import xarray as xr
holoviews.config.image_rtol = 1e-2
auth = earthaccess.login()
2. Search for L3M PACE Trace Gas Data#
Level-2 (L2) and Level-3 mapped (L3M) trace gas (TRGAS) data products are available on NASA Earthdata.
Let’s search and open up the first L3M file collected during the temporal span of January 2025 to December 2025:
tspan = ("2025-01-01", "2025-12-31")
results = earthaccess.search_data(
short_name="PACE_OCI_L3M_TRGAS",
temporal=tspan,
count=1,
)
paths = earthaccess.open(results)
paths
[<File-like object S3FileSystem, ob-cumulus-prod-public/PACE_OCI.20250101_20250131.L3m.MO.TRGAS.V3_0.0p1deg.nc>]
You can see that this search opened PACE_OCI.20250101_20250131.L3m.MO.TRGAS.V3_0.0p1deg.nc which is a L3M monthly (MO) composite with a 0.1° (0p1deg) pixel resolution.”
3. Open as an XArray dataset#
We will use xarray.open_dataset to open the NetCDF and then merge them into a single dataset.
dataset = xr.open_dataset(paths[0])
dataset
<xarray.Dataset> Size: 52MB
Dimensions: (lat: 1800, lon: 3600)
Coordinates:
* lat (lat) float32 7kB 89.95 89.85 89.75 ... -89.85 -89.95
* lon (lon) float32 14kB -179.9 -179.9 -179.8 ... 179.9 180.0
Data variables:
total_column_o3 (lat, lon) float32 26MB ...
total_column_no2 (lat, lon) float32 26MB ...
Attributes: (12/54)
product_name: PACE_OCI.20250101_20250131.L3m.MO.TRGA...
instrument: OCI
title: OCI Level-3 Standard Mapped Image
project: Ocean Biology Processing Group (NASA/G...
platform: PACE
source: satellite observations from OCI-PACE
... ...
keywords:
id: L3/PACE_OCI.20250101_20250131.L3m.MO.T...
history: l3mapmerge.py -v l3mapmerge_inputs.txt...
processing_version: 3.0
identifier_product_doi_authority: https://dx.doi.org
identifier_product_doi: 10.5067/PACE/OCI/L3M/TRGAS/3.0If you expand the total_column_no2 variable, you’ll see that it is a 2D variable consisting of total vertical column NO2 measurements with units of molecules cm-2.
Let’s plot it!
fig, ax = plt.subplots(figsize=(9, 5), subplot_kw={"projection": ccrs.PlateCarree()})
ax.coastlines(linewidth=0.2)
ax.add_feature(cfeature.BORDERS, linewidth=0.1)
ax.add_feature(cfeature.OCEAN, linewidth=0.2)
ax.add_feature(cfeature.LAKES, linewidth=0.2)
ax.add_feature(cfeature.LAND, facecolor="oldlace", linewidth=0.2)
cmap = matplotlib.colors.LinearSegmentedColormap.from_list(
name="no2",
colors=["lightgrey", "cyan", "yellow", "orange", "red", "darkred"],
)
dataset["total_column_no2"].plot(x="lon", y="lat", vmin=3e15, vmax=10e15, cmap=cmap)
ax.gridlines(
draw_labels={"left": "y", "bottom": "x"},
linewidth=0.25,
color="grey",
alpha=0.8,
)
plt.show()
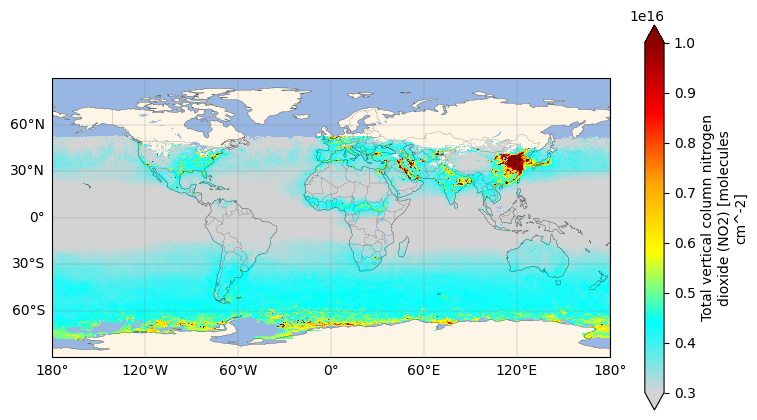
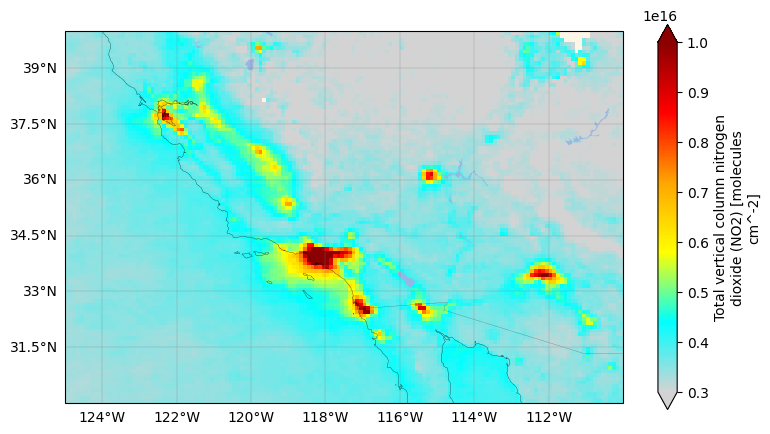
Let’s zoom in to Los Angeles, California.
plt.sca(ax)
plt.xlim(-125, -110)
plt.ylim(30, 40)
plt.show()
4. Interactive NO2 plot#
An interative plot allows you to engage with the data such as zooming, panning, and hovering over for more information. We will use the hvplot accessor on XArray data structures to make an interactive plot of the single file we accessed above.
dataset["total_column_no2"].hvplot(
x="lon",
y="lat",
cmap=cmap,
clim=(3e15, 10e15),
aspect=2,
title="Total vertical column NO₂ April 2024",
widget_location="top",
geo=True,
coastline=True,
tiles="EsriWorldStreetMap",
)
