# Install external dependencies
!pip install -qq netcdf4 xarray harmony-py python-cmr geopandas wgetDemonstration for working with TEMPO data via Harmony-py
This notebook is based on examples from the harmony-py repository and https://harmony.earthdata.nasa.gov/docs. - Created: 3 May 2023 - Last updated: 9 Apr 2024
What is Harmony?
Harmony is the cloud services orchestrator for NASA Earth Science Data and Information System (ESDIS). The goal of Harmony is to provide services to increase usage and ease of use of ESDIS’ Earth observation data, especially focusing on opportunities made possible by cloud-accessible data.
Data processed by Harmony are staged in Amazon s3 buckets, and Harmony services run in containers in pods in a Kubernetes cluster.
Note that services provided via Harmony can transform NASA data as well as provide access to the original data, through an Application Programmable Interface (API)…
Harmony services can be invoked through curl commands, e.g.:
https://harmony.earthdata.nasa.gov/{collectionId}/ogc-api-coverages/1.0.0/{variable}/coverage/rangeset
(Harmony services REST API conforms to the OGC Coverages API version 1.0.0. It accepts parameters in the URL path as well as query parameters.)
Harmony can also be invoked via wrapper libraries such as harmony-py, which is demonstrated below.
Set-up
# Load packages into current runtime
import datetime as dt
import getpass
import traceback
from typing import Dict
import numpy as np
import numpy.ma as ma
import netCDF4 as nc
import xarray as xr
import geopandas as gpd
import matplotlib.pyplot as plt
from matplotlib.pyplot import cm
import cmr
from harmony import BBox, Client, Collection, Request
from harmony.config import Environment# Get United States shapefile as a graphics base image
!wget -q https://www2.census.gov/geo/tiger/GENZ2018/shp/cb_2018_us_state_20m.zip
!unzip cb_2018_us_state_20m.zip
!rm cb_2018_us_state_20m.zip
gdf_states = gpd.read_file("cb_2018_us_state_20m.shp")Archive: cb_2018_us_state_20m.zip
inflating: cb_2018_us_state_20m.shp.ea.iso.xml
inflating: cb_2018_us_state_20m.shp.iso.xml
inflating: cb_2018_us_state_20m.shp
inflating: cb_2018_us_state_20m.shx
inflating: cb_2018_us_state_20m.dbf
inflating: cb_2018_us_state_20m.prj
extracting: cb_2018_us_state_20m.cpg Retrieve a data file (via Harmony)
All users will need an Earthdata Login (EDL) account in order to access NASA data and services.
Once a user has an EDL username and password they will need to use these when accessing Harmony.
print('Please provide your Earthdata Login credentials to allow data access')
print('Your credentials will only be passed to Earthdata and will not be exposed in the notebook')
username = input('Username:')
harmony_client = Client(env=Environment.PROD, auth=(username, getpass.getpass()))Please provide your Earthdata Login credentials to allow data access
Your credentials will only be passed to Earthdata and will not be exposed in the notebookUsername: dkaufas
········Choose a granule, and write out the request with a chosen collection ID and granule name.
In this case, we’ll choose a granule (a ~6 minute window) containing nitrogen dioxide data crossing part of the great lakes region.
# "Nitrogen Dioxide total column"
request = Request(
collection=Collection(id='C2724057189-LARC_CLOUD'),
# granule_name=['TEMPO_NO2_L2_V01_20231230T232423Z_S011G06.nc']
granule_name=["TEMPO_NO2_L2_V01_20231230T152429Z_S004G03.nc"]
)
request.is_valid()True# Submit the request
job_id = harmony_client.submit(request)
print(f'jobID = {job_id}')
# Wait for the processing to complete.
harmony_client.wait_for_processing(job_id, show_progress=True)jobID = e18a62ca-c485-4496-b422-a7b2794d31d7 [ Processing: 100% ] |###################################################| [|]# Download the resulting files
results = harmony_client.download_all(job_id, directory='/tmp', overwrite=True)
all_results_stored = [f.result() for f in results]
print(f"Number of result files: {len(all_results_stored)}")Number of result files: 1Open the data file
# Open the data file, in this case using the Xarray package
ds_root = xr.open_dataset(all_results_stored[0])
ds_geo = xr.open_dataset(all_results_stored[0], group='geolocation')
ds_product = xr.open_dataset(all_results_stored[0], group='product')
# Merge the groups since Xarray doesn't yet natively combine groups for netCDF4
ds = xr.merge([ds_root, ds_geo, ds_product])
ds<xarray.Dataset> Size: 28MB
Dimensions: (xtrack: 2048, mirror_step: 131,
corner: 4)
Coordinates:
* xtrack (xtrack) int32 8kB 0 1 ... 2047
* mirror_step (mirror_step) int32 524B 264 ......
time (mirror_step) datetime64[ns] 1kB ...
latitude (mirror_step, xtrack) float32 1MB ...
longitude (mirror_step, xtrack) float32 1MB ...
Dimensions without coordinates: corner
Data variables: (12/13)
latitude_bounds (mirror_step, xtrack, corner) float32 4MB ...
longitude_bounds (mirror_step, xtrack, corner) float32 4MB ...
solar_zenith_angle (mirror_step, xtrack) float32 1MB ...
solar_azimuth_angle (mirror_step, xtrack) float32 1MB ...
viewing_zenith_angle (mirror_step, xtrack) float32 1MB ...
viewing_azimuth_angle (mirror_step, xtrack) float32 1MB ...
... ...
vertical_column_total (mirror_step, xtrack) float64 2MB ...
vertical_column_total_uncertainty (mirror_step, xtrack) float64 2MB ...
main_data_quality_flag (mirror_step, xtrack) float32 1MB ...
vertical_column_troposphere (mirror_step, xtrack) float64 2MB ...
vertical_column_stratosphere (mirror_step, xtrack) float64 2MB ...
vertical_column_troposphere_uncertainty (mirror_step, xtrack) float64 2MB ...
Attributes: (12/36)
tio_commit: abba4bbcf910f6b8213ce2dfcabad202a0152ea9
product_type: NO2
processing_level: 2
processing_version: 1
scan_num: 4
granule_num: 3
... ...
collection_shortname: TEMPO_NO2_L2
collection_version: 1
keywords: EARTH SCIENCE>ATMOSPHERE>AIR QUALITY>NI...
summary: Nitrogen dioxide Level 2 files provide ...
coremetadata: \nGROUP = INVENTORYMET...
history: 2023-12-30T19:15:14Z:/tempo/nas0/sdpc_s...Visualize one of the data variables
Here we take a look at the Vertical Column Total variable, both its metadata and then by creating a map visualization. Note the use of main_data_quality_flag in the plotting code to ensure that we are examining only the “normal” quality data.
product_variable_name = 'vertical_column_total'
ds[product_variable_name]<xarray.DataArray 'vertical_column_total' (mirror_step: 131, xtrack: 2048)> Size: 2MB
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]])
Coordinates:
* xtrack (xtrack) int32 8kB 0 1 2 3 4 5 ... 2043 2044 2045 2046 2047
* mirror_step (mirror_step) int32 524B 264 265 266 267 ... 391 392 393 394
time (mirror_step) datetime64[ns] 1kB 2023-12-30T15:24:47.8762749...
latitude (mirror_step, xtrack) float32 1MB 58.76 58.7 ... 17.25 17.24
longitude (mirror_step, xtrack) float32 1MB -63.82 -63.87 ... -83.69
Attributes:
long_name: nitrogen dioxide vertical column
comment: nitrogen dioxide vertical column determined from fitted slant...
units: molecules/cm^2fig, ax = plt.subplots(figsize=(10, 6))
gdf_states.plot(ax=ax, color='gray')
contour_handle = (
ds[product_variable_name]
.where(ds["main_data_quality_flag"] == 0)
.plot
.contourf(ax=ax, x='longitude', y='latitude', levels=100, vmin=0)
)
ax.set_xlim((-150, -40))
ax.set_ylim((14, 65))
plt.show()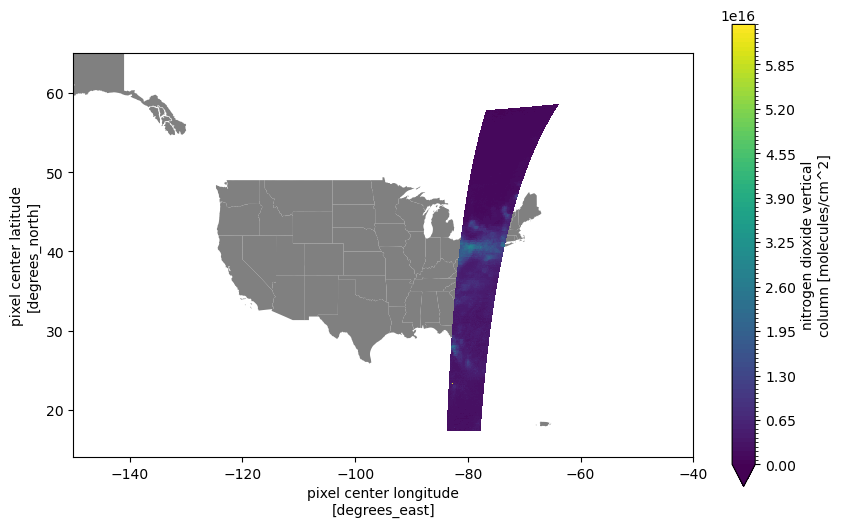
Zonal means
# define two-degree wide latitude bins
lat_bins = np.arange(15, 61, 5)
# define a label for each bin corresponding to the central latitude
lat_center = np.arange(15, 60, 5)
# group according to those bins and take the mean
product_lat_mean = (ds[product_variable_name]
.groupby_bins("latitude", lat_bins, labels=lat_center)
.mean(dim=xr.ALL_DIMS)
)
product_lat_mean.plot()
plt.gca().invert_xaxis()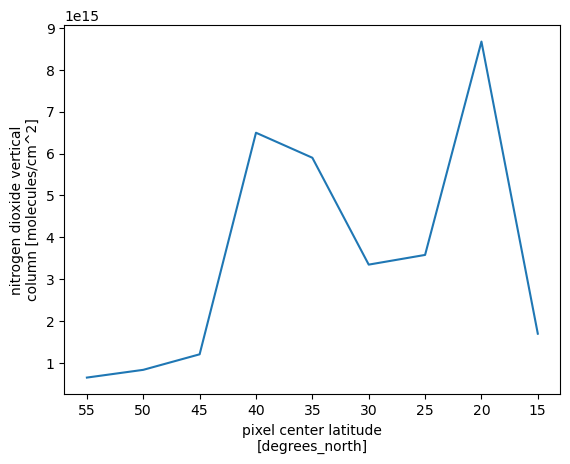
(ds_product[product_variable_name]
.where(ds["main_data_quality_flag"] == 0)
.plot
.contourf(x='mirror_step', y='xtrack', vmin=0)
)
plt.gca().invert_xaxis()
plt.show()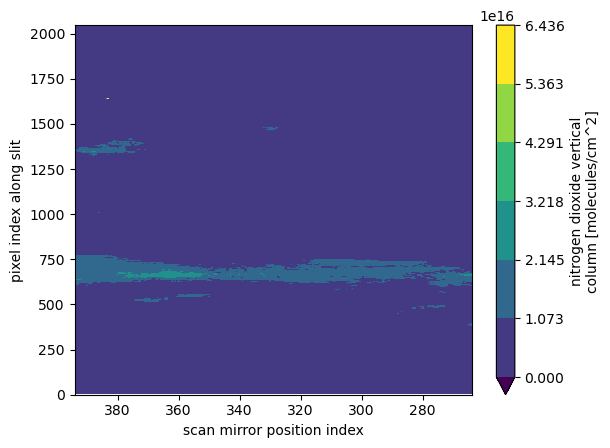
Retrieve only a single variable
# TEMPO Formaldehyde
request = Request(
collection=Collection(id='C2732717000-LARC_CLOUD'),
granule_name=['TEMPO_HCHO_L2_V01_20231230T145736Z_S003G08.nc'],
variables=['product/vertical_column']
)
job_id = harmony_client.submit(request)
print(f'jobID = {job_id}')
harmony_client.wait_for_processing(job_id, show_progress=True)
# Download the resulting files
results = harmony_client.download_all(job_id, directory='/tmp', overwrite=True)
all_results_stored = [f.result() for f in results]
print(f"Number of result files: {len(all_results_stored)}")jobID = 2a23003a-414e-4802-8905-117badc91749 [ Processing: 100% ] |###################################################| [|]Number of result files: 1# Open the data file, in this case using the Xarray package
ds_root = xr.open_dataset(all_results_stored[0])
ds_geo = xr.open_dataset(all_results_stored[0], group='geolocation')
ds_product = xr.open_dataset(all_results_stored[0], group='product')
# Merge the groups since Xarray doesn't yet natively combine groups for netCDF4
ds = xr.merge([ds_root, ds_geo, ds_product])
ds<xarray.Dataset> Size: 4MB
Dimensions: (mirror_step: 131, xtrack: 2048)
Dimensions without coordinates: mirror_step, xtrack
Data variables:
time (mirror_step) datetime64[ns] 1kB ...
latitude (mirror_step, xtrack) float32 1MB ...
longitude (mirror_step, xtrack) float32 1MB ...
vertical_column (mirror_step, xtrack) float64 2MB ...
Attributes: (12/37)
tio_commit: abba4bbcf910f6b8213ce2dfcabad202a0152ea9
product_type: HCHO
processing_level: 2
processing_version: 1
scan_num: 3
granule_num: 8
... ...
collection_version: 1
keywords: EARTH SCIENCE>ATMOSPHERE>AIR QUALITY>VO...
summary: Formaldehyde Level 2 files provide trac...
coremetadata: \nGROUP = INVENTORYMET...
history: 2023-12-30T18:08:18Z:/tempo/nas0/sdpc_s...
history_json: [{"date_time": "2024-04-09T16:08:53.010...Note the reduced list of data variables.
Retrieve data from a select time range
#@title
# A clean-up step to ensure there isn't a clash between newly downloaded granules
for rf in all_results_stored:
!rm {rf}request = Request(collection=Collection(id='C2732717000-LARC_CLOUD'),
# Note there is not a granule specified!
temporal={
'start': dt.datetime(2023, 12, 30, 22, 30, 0),
'stop': dt.datetime(2023, 12, 30, 22, 45, 0)
})
job_id = harmony_client.submit(request)
print(f'jobID = {job_id}')
harmony_client.wait_for_processing(job_id, show_progress=True)
# Download the resulting files
results = harmony_client.download_all(job_id, directory='/tmp', overwrite=True)
all_results_stored = [f.result() for f in results]
print(f"Number of result files: {len(all_results_stored)}")jobID = deaeb382-93eb-472e-a9ad-ad1f6175a14f [ Processing: 100% ] |###################################################| [|]Number of result files: 4# Open the data files
ds_dict = dict()
for r in sorted(all_results_stored):
ds_root = xr.open_dataset(r)
ds_geo = xr.open_dataset(r, group='geolocation')
ds_product = xr.open_dataset(r, group='product')
ds_dict[r] = xr.merge([ds_root, ds_geo, ds_product])
print(f"Time range: {ds_dict[r]['time'].values.min()} - {ds_dict[r]['time'].values.max()}")Time range: 2023-12-30T22:30:00.279620864 - 2023-12-30T22:31:19.139312896
Time range: 2023-12-30T22:31:22.172382976 - 2023-12-30T22:37:56.470801152
Time range: 2023-12-30T22:37:59.503866880 - 2023-12-30T22:44:33.802316800
Time range: 2023-12-30T22:44:36.835376128 - 2023-12-30T22:44:58.066825984Note how the time ranges fit within the requested temporal range
# Visualize each data file
fig, axs = plt.subplots(nrows=len(ds_dict), ncols=1,
sharex=True, figsize=(7, 10),
gridspec_kw=dict(hspace=0.25))
for i, (dk, dv) in enumerate(ds_dict.items()):
Var = dv['vertical_column']
ax = axs[i]
if np.count_nonzero(~np.isnan(Var.values)) > 0:
gdf_states.plot(ax=ax, color='gray')
Var.plot.contourf(ax=ax, x='longitude', y='latitude', levels=100, vmin=0)
ax.set_xlim((-150, -40))
ax.set_ylim((14, 65))
ax.set_title(dk, fontsize=8)
plt.show()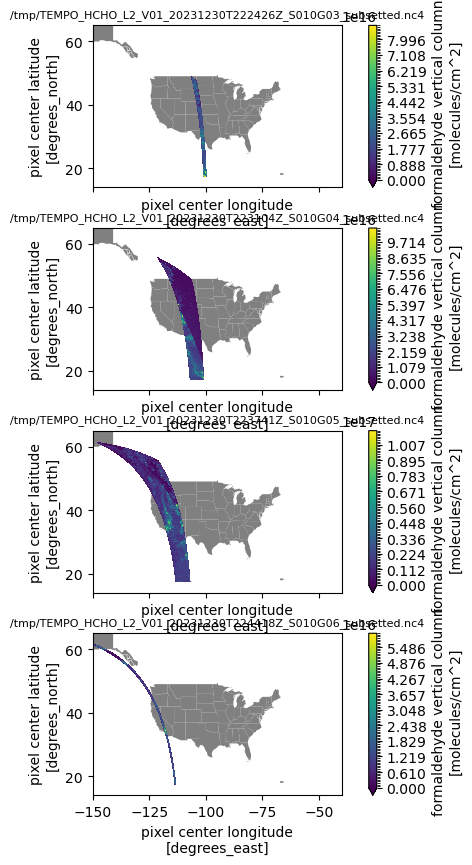
# A clean-up step to ensure there isn't a clash between newly downloaded granules
for rf in all_results_stored:
!rm {rf}Retrieve data from a spatial bounding box
request = Request(collection=Collection(id='C2732717000-LARC_CLOUD'),
# Note there is not a granule specified!
spatial=BBox(-115, 35, -95, 45),
temporal={
'start': dt.datetime(2023, 12, 30, 22, 30, 0),
'stop': dt.datetime(2023, 12, 30, 22, 45, 0)
})
job_id = harmony_client.submit(request)
print(f'jobID = {job_id}')
harmony_client.wait_for_processing(job_id, show_progress=True)
# Download the resulting files
results = harmony_client.download_all(job_id, directory='/tmp', overwrite=True)
all_results_stored = [f.result() for f in results]
print(f"Number of result files: {len(all_results_stored)}")jobID = 03cd6c83-c1cf-46b2-b64d-7e5f37573c05 [ Processing: 100% ] |###################################################| [|]Number of result files: 3# Open the data files
ds_dict = dict()
for r in sorted(all_results_stored):
ds_root = xr.open_dataset(r)
ds_geo = xr.open_dataset(r, group='geolocation')
ds_product = xr.open_dataset(r, group='product')
ds_dict[r] = xr.merge([ds_root, ds_geo, ds_product])# ds_dict['/tmp/TEMPO_NO2-PROXY_L2_V01_20140430T222959Z_S012G06_subsetted.nc4']['vertical_column_total']# Visualize each data file
fig, axs = plt.subplots(nrows=len(ds_dict), ncols=1,
sharex=True, figsize=(7, 10),
gridspec_kw=dict(hspace=0.1))
for i, (dk, dv) in enumerate(ds_dict.items()):
Var = dv['vertical_column']
ax = axs[i]
if np.count_nonzero(~np.isnan(Var.values)) > 0:
# Zm = ma.masked_where(np.isnan(Var), Var)
# Var[:] = Zm
# Var.plot(x='longitude', y='latitude', ax=axs[i])
gdf_states.plot(ax=ax, color='gray')
ax.scatter(dv['longitude'], dv['latitude'], s=1, c=dv['vertical_column'])
ax.set_title(dk, fontsize=8)
ax.set_xlim((-150, -40))
ax.set_ylim((14, 65))
ax.grid(color='gray', linestyle='--', linewidth=0.5)
# Coordinates of rectangle vertices in clockwise order
xs = [-115, -115, -95, -95, -115]
ys = [35, 45, 45, 35, 35]
ax.plot(xs, ys, color="red", linestyle=':')
plt.show()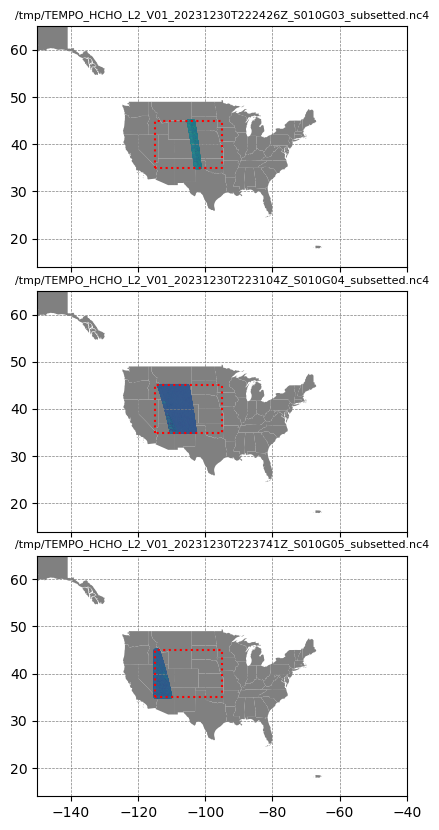
Note the cut-off of data for the bounding box: (-115, 35, -95, 45)
End of Notebook.
