import json
import os
import re
import time
from datetime import datetime
from glob import glob
from pathlib import Path
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import dask # not called directly, but necessary for using xarray open_mfdataset()
import netCDF4 as nc
import numpy as np
import xarray as xr
import matplotlib.pyplot as plt
from numpy import ma
from tqdm.notebook import tqdm
import earthaccess
# For the timeseries section
import json
from scipy import spatial
# For the timeseries section, to compare with EPA Air Quality data
import pyrsig
# Fot the animation section
import matplotlib.animation as animation
from matplotlib.animation import FuncAnimation, PillowWriterExploring data from DSCOVR EPIC Level 2 Aerosol Version 3
Note that operations that save the figures are commented out, and so will need to be uncommented in order to save new figures.
- Created: 2023 August 29
- Last Modified: 2024 October 28
- Author: Daniel Kaufman daniel.kaufman@nasa.gov
Overview
This notebook visualizes the spatial transport and temporal variations of UV Aerosol Index (UVAI) from DSCOVR EPIC, and compares those measurements to EPA air quality data for the same time period. The figures from this notebook were included in the poster “Impact of Canadian Wildfires on Mid Atlantic’s Region Air Quality: An Analysis Using ASDC Data” (A41O-2864) presented at the 2023 American Geophysical Union (AGU) Fall Meeting.
Table of Contents
- Setup
- Downloads DSCOVR EPIC data files
- Converts the data to Zarr stores
- Makes map graphic of a single time
- Makes multi-panel figure
- Makes timeseries figure that compares to EPA air quality data
- Creates an animation
Dataset Information
“DSCOVR_EPIC_L2_AER_03 is the Deep Space Climate Observatory (DSCOVR) Enhanced Polychromatic Imaging Camera (EPIC) Level 2 UV Aerosol Version 3 data product. Observations for this data product are at 340 and 388 nm and are used to derive near UV (ultraviolet) aerosol properties. The EPIC aerosol retrieval algorithm (EPICAERUV) uses a set of aerosol models to account for the presence of carbonaceous aerosols from biomass burning and wildfires (BIO), desert dust (DST), and sulfate-based (SLF) aerosols. These aerosol models are identical to those assumed in the OMI (Ozone Monitoring Instrument) algorithm (Torres et al., 2007; Jethva and Torres, 2011).” (Source)
This requires the data granules from DSCOVR EPIC Level-2 to have been downloaded into a local directory.
1. Setup
# 2. Download data files
In this section, we use earthaccess and free Earthdata credentials to login to Earthdata, find the data granules corresponding to our chosen time and spatial bounds, and then download the files.
auth = earthaccess.login()
# are we authenticated?
if not auth.authenticated:
# ask for credentials and persist them in a .netrc file
auth.login(strategy="interactive", persist=True)collection_id = 'C1962643459-LARC_ASDC'
# Bounds within which we search for data granules
date_start = "2023-06-01 00:00"
date_end = "2023-06-16 23:59"
date_range = (date_start, date_end)
bbox = (-142, 31, -49, 73) # min lon, min lat, max lon, max lat
# Find the data granules and their links
results = earthaccess.search_data(
concept_id = collection_id,
temporal = date_range,
bounding_box = bbox,
)Granules found: 336Modify the following cell to specify the local directory where the data granules (will) reside.
data_dir = Path("~/data/dscovr").resolve()!! Warning !! This next cell can take a long time. It needs to be run once, but then should be commented-out if the data files are already in a local directory.
# # Download the files
# downloaded_files = earthaccess.download(
# results,
# local_path=data_dir,
# )Get the list of files that are present
pattern = re.compile(r"DSCOVR_EPIC_L2_AER_03_([0-9]+)_03") # the capture group is a datetime timestamp
file_list = list(data_dir.glob("DSCOVR_EPIC_L2_AER_*.he5"))
num_files = len(file_list)# These are the paths to groups *within* each HDF file
data_fields_path = 'HDFEOS/SWATHS/Aerosol NearUV Swath/Data Fields'
geolocations_path = 'HDFEOS/SWATHS/Aerosol NearUV Swath/Geolocation Fields'3. Wrangle data into Zarr stores
In this section, the HDF files are converted to Zarr stores to enable faster access for data manipulation later in the notebook. And then the dataset is opened.
def convert_hdf_to_xrds(filepath: Path, group_path: str) -> xr.Dataset:
"""Converts a HDF group into an xarray Dataset with an added time dimension
Parameters
----------
filepath : pathlib.Path
The path to a DSCOVR EPIC HDF file
group_path : str
The internal path to a group, e.g., 'HDFEOS/SWATHS/Aerosol NearUV Swath/Data Fields'
"""
# The filename's datetime string is converted into a datetime timestamp
timestamp = pattern.findall(str(fp.stem))[0]
timestamp_dt = datetime.strptime(timestamp, "%Y%m%d%H%M%S")
# Note: the netCDF library works to access these datasets, so we just use it instead of h5py.
with nc.Dataset(filepath) as ds:
grp = ds[group_path]
# The HDF group is converted into an xarray Dataset object, and then
# a new singleton 'time' dimension is added to the Dataset with the timestamp as its only value.
grp_ds = (
xr.open_dataset(xr.backends.NetCDF4DataStore(grp))
.expand_dims(time=[timestamp_dt], axis=0)
)
return grp_ds!! Warning !! This next cell can take a long time (>=30 min.). It needs to be run once, but then should be commented-out if the zarr files already exist.
print("Extracting groups...")
idcs_to_do = range(0, len(file_list))
for idx in tqdm(idcs_to_do, total=len(idcs_to_do)):
new_path = data_dir / f"dscovr_zarrstore_{idx:03}.zarr"
if not new_path.exists():
fp = file_list[idx]
(
convert_hdf_to_xrds(fp, data_fields_path)
.rename({"phony_dim_0": "XDim",
"phony_dim_1": "YDim",
"phony_dim_2": "nLayers",
"phony_dim_3": "nWave3" ,
"phony_dim_4": "nWave2"})
).merge(
(convert_hdf_to_xrds(fp, geolocations_path)
.rename({"phony_dim_5": "XDim",
"phony_dim_6": "YDim"})
)
).to_zarr(new_path, consolidated=True)
# if idx > 2:
# break
print("Done extracting groups.")Now let’s open the dataset. This may take around 30 seconds or so.
mfds = xr.open_mfdataset(data_dir.glob("dscovr_zarrstore_*.zarr"),
engine='zarr', combine='by_coords')
mfds<xarray.Dataset> Size: 624GB
Dimensions: (time: 341, XDim: 2048, YDim: 2048,
nLayers: 5, nWave3: 3, nWave2: 2)
Coordinates:
* time (time) datetime64[ns] 3kB 2023-06-01T00:0...
Dimensions without coordinates: XDim, YDim, nLayers, nWave3, nWave2
Data variables: (12/36)
AIRSCO_Flags (time, XDim, YDim) float32 6GB dask.array<chunksize=(1, 512, 1024), meta=np.ndarray>
AIRSL3COvalue (time, XDim, YDim) float32 6GB dask.array<chunksize=(1, 256, 512), meta=np.ndarray>
AerosolAbsOpticalDepthVsHeight (time, XDim, YDim, nLayers, nWave3) float32 86GB dask.array<chunksize=(1, 256, 512, 2, 1), meta=np.ndarray>
AerosolCorrCloudOpticalDepth (time, XDim, YDim) float32 6GB dask.array<chunksize=(1, 256, 512), meta=np.ndarray>
AerosolOpticalDepthOverCloud (time, XDim, YDim, nWave3) float32 17GB dask.array<chunksize=(1, 512, 512, 1), meta=np.ndarray>
AerosolOpticalDepthVsHeight (time, XDim, YDim, nLayers, nWave3) float32 86GB dask.array<chunksize=(1, 256, 512, 2, 1), meta=np.ndarray>
... ...
SurfaceAlbedo (time, XDim, YDim, nWave2) float32 11GB dask.array<chunksize=(1, 512, 512, 1), meta=np.ndarray>
SurfaceType (time, XDim, YDim) float32 6GB dask.array<chunksize=(1, 512, 512), meta=np.ndarray>
TerrainPressure (time, XDim, YDim) float32 6GB dask.array<chunksize=(1, 256, 512), meta=np.ndarray>
UVAerosolIndex (time, XDim, YDim) float32 6GB dask.array<chunksize=(1, 256, 512), meta=np.ndarray>
ViewingZenithAngle (time, XDim, YDim) float32 6GB dask.array<chunksize=(1, 256, 512), meta=np.ndarray>
Wavelength (time, nWave3) float32 4kB dask.array<chunksize=(1, 3), meta=np.ndarray>mfds['Longitude'].shape(341, 2048, 2048)Prepare for static figures
In this subsection, we prepare a color scheme, choose the data variable and spatial domain of interest, and define a couple utility functions for use later in the notebook.
- The color scheme is created by slightly modifying the “turbo” colormap for improved visualization.
from matplotlib.colors import ListedColormap
from matplotlib import colormaps
# Add a white 'under' color to the turbo colormap
turbo_cm = colormaps.get_cmap('turbo')
newcolors = turbo_cm(np.linspace(0, 1, 256))
silver_color = np.array([0.95, 0.95, 0.95, 1])
newcolors[0, :] = silver_color
my_turbo = ListedColormap(newcolors)
my_turbo- The data variable chosen here will be used for all the figures to follow.
# Choose the data variable here.
data_variable = "UVAerosolIndex"- The spatial domain defined here will determine the visualizations to follow.
extent = [-141, -50, 32, 72]
central_lon = np.mean(extent[:2])
central_lat = np.mean(extent[2:])- These utility functions will simplify some code blocks later.
def get_geo_mask(lon, lat):
"""This masks given lon/lat arrays to the chosen spatial domain."""
lon_mask = (lon > extent[0]) & (lon < extent[1])
lat_mask = (lat > extent[2]) & (lat < extent[3])
return lon_mask & lat_mask
def get_data_arrays(a_timestep: int):
"""Given a chose timestep, this extracts data arrays corresponding to that time."""
geo_mask = get_geo_mask(mfds['Longitude'][a_timestep, :, :], mfds['Latitude'][a_timestep, :, :]).compute().values
lon_values = mfds['Longitude'][a_timestep, :, :].where(geo_mask).values
lat_values = mfds['Latitude'][a_timestep, :, :].where(geo_mask).values
var_values = mfds[data_variable][a_timestep, :, :].where(geo_mask).values
timestamp = datetime.strptime(
np.datetime_as_string(mfds['time'][a_timestep].values, unit='ms'),
"%Y-%m-%dT%H:%M:%S.%f"
)
# Replace any +inf or -inf values with NaN.
var_values[np.isinf(var_values)] = np.nan
return lon_values, lat_values, var_values, timestamp# 4. Make map of a single timestep
Now let’s pick a timestep to visualize. First print out the timestep indices and their values so we can choose one.
for i, x in enumerate(mfds['time']):
print(f"{i} -- {x.values}")0 -- 2023-06-01T00:08:31.000000000
1 -- 2023-06-01T01:13:58.000000000
2 -- 2023-06-01T02:19:25.000000000
3 -- 2023-06-01T03:24:52.000000000
4 -- 2023-06-01T04:30:19.000000000
5 -- 2023-06-01T05:35:46.000000000
6 -- 2023-06-01T06:41:13.000000000
7 -- 2023-06-01T07:46:40.000000000
8 -- 2023-06-01T08:52:08.000000000
9 -- 2023-06-01T09:57:35.000000000
10 -- 2023-06-01T11:03:02.000000000
11 -- 2023-06-01T12:08:30.000000000
12 -- 2023-06-01T13:13:57.000000000
13 -- 2023-06-01T14:19:24.000000000
14 -- 2023-06-01T15:24:51.000000000
15 -- 2023-06-01T16:30:19.000000000
16 -- 2023-06-01T17:35:46.000000000
17 -- 2023-06-01T18:41:13.000000000
18 -- 2023-06-01T19:46:40.000000000
19 -- 2023-06-01T20:52:08.000000000
20 -- 2023-06-01T21:57:35.000000000
21 -- 2023-06-01T23:03:03.000000000
22 -- 2023-06-02T00:27:13.000000000
23 -- 2023-06-02T01:32:40.000000000
24 -- 2023-06-02T02:38:07.000000000
25 -- 2023-06-02T03:43:34.000000000
26 -- 2023-06-02T04:49:01.000000000
27 -- 2023-06-02T05:54:28.000000000
28 -- 2023-06-02T06:59:56.000000000
29 -- 2023-06-02T08:05:23.000000000
30 -- 2023-06-02T09:10:50.000000000
31 -- 2023-06-02T10:16:17.000000000
32 -- 2023-06-02T11:21:44.000000000
33 -- 2023-06-02T12:27:12.000000000
34 -- 2023-06-02T13:32:39.000000000
35 -- 2023-06-02T14:38:07.000000000
36 -- 2023-06-02T15:43:34.000000000
37 -- 2023-06-02T16:49:02.000000000
38 -- 2023-06-02T17:54:29.000000000
39 -- 2023-06-02T18:59:57.000000000
40 -- 2023-06-02T20:05:24.000000000
41 -- 2023-06-02T21:10:52.000000000
42 -- 2023-06-02T22:16:19.000000000
43 -- 2023-06-02T23:21:46.000000000
44 -- 2023-06-03T00:45:54.000000000
45 -- 2023-06-03T01:51:22.000000000
46 -- 2023-06-03T02:56:49.000000000
47 -- 2023-06-03T04:02:17.000000000
48 -- 2023-06-03T05:07:44.000000000
49 -- 2023-06-03T06:13:11.000000000
50 -- 2023-06-03T07:18:39.000000000
51 -- 2023-06-03T17:07:45.000000000
52 -- 2023-06-03T18:13:12.000000000
53 -- 2023-06-03T19:18:39.000000000
54 -- 2023-06-03T20:24:06.000000000
55 -- 2023-06-03T21:29:34.000000000
56 -- 2023-06-03T22:35:01.000000000
57 -- 2023-06-03T23:40:28.000000000
58 -- 2023-06-04T01:04:37.000000000
59 -- 2023-06-04T02:10:04.000000000
60 -- 2023-06-04T03:15:31.000000000
61 -- 2023-06-04T04:20:58.000000000
62 -- 2023-06-04T05:26:25.000000000
63 -- 2023-06-04T06:31:53.000000000
64 -- 2023-06-04T07:37:20.000000000
65 -- 2023-06-04T08:42:47.000000000
66 -- 2023-06-04T09:48:14.000000000
67 -- 2023-06-04T10:53:42.000000000
68 -- 2023-06-04T11:59:09.000000000
69 -- 2023-06-04T13:04:36.000000000
70 -- 2023-06-04T14:10:04.000000000
71 -- 2023-06-04T15:15:31.000000000
72 -- 2023-06-04T16:20:58.000000000
73 -- 2023-06-04T17:26:26.000000000
74 -- 2023-06-04T18:31:53.000000000
75 -- 2023-06-04T19:37:20.000000000
76 -- 2023-06-04T20:42:47.000000000
77 -- 2023-06-04T21:48:15.000000000
78 -- 2023-06-04T22:53:42.000000000
79 -- 2023-06-05T00:55:16.000000000
80 -- 2023-06-05T02:00:43.000000000
81 -- 2023-06-05T03:06:10.000000000
82 -- 2023-06-05T04:11:37.000000000
83 -- 2023-06-05T05:17:05.000000000
84 -- 2023-06-05T06:22:32.000000000
85 -- 2023-06-05T08:33:26.000000000
86 -- 2023-06-05T09:38:54.000000000
87 -- 2023-06-05T10:44:21.000000000
88 -- 2023-06-05T11:49:49.000000000
89 -- 2023-06-05T12:55:16.000000000
90 -- 2023-06-05T14:00:43.000000000
91 -- 2023-06-05T15:06:10.000000000
92 -- 2023-06-05T16:11:37.000000000
93 -- 2023-06-05T18:22:32.000000000
94 -- 2023-06-05T19:28:00.000000000
95 -- 2023-06-05T20:33:27.000000000
96 -- 2023-06-05T21:38:55.000000000
97 -- 2023-06-05T22:44:21.000000000
98 -- 2023-06-05T23:49:49.000000000
99 -- 2023-06-06T00:36:34.000000000
100 -- 2023-06-06T01:42:01.000000000
101 -- 2023-06-06T02:47:28.000000000
102 -- 2023-06-06T03:52:55.000000000
103 -- 2023-06-06T03:53:48.000000000
104 -- 2023-06-06T04:58:22.000000000
105 -- 2023-06-06T06:03:49.000000000
106 -- 2023-06-06T07:09:16.000000000
107 -- 2023-06-06T08:14:44.000000000
108 -- 2023-06-06T09:20:11.000000000
109 -- 2023-06-06T10:25:38.000000000
110 -- 2023-06-06T11:31:05.000000000
111 -- 2023-06-06T12:36:32.000000000
112 -- 2023-06-06T13:42:00.000000000
113 -- 2023-06-06T14:47:27.000000000
114 -- 2023-06-06T15:52:54.000000000
115 -- 2023-06-06T16:58:21.000000000
116 -- 2023-06-06T18:03:49.000000000
117 -- 2023-06-06T19:09:16.000000000
118 -- 2023-06-06T20:14:43.000000000
119 -- 2023-06-06T21:20:11.000000000
120 -- 2023-06-06T22:25:37.000000000
121 -- 2023-06-06T23:31:05.000000000
122 -- 2023-06-07T00:17:51.000000000
123 -- 2023-06-07T01:23:19.000000000
124 -- 2023-06-07T02:28:46.000000000
125 -- 2023-06-07T03:34:13.000000000
126 -- 2023-06-07T04:39:40.000000000
127 -- 2023-06-07T05:45:07.000000000
128 -- 2023-06-07T06:50:35.000000000
129 -- 2023-06-07T07:56:02.000000000
130 -- 2023-06-07T09:01:30.000000000
131 -- 2023-06-07T10:06:57.000000000
132 -- 2023-06-07T11:12:24.000000000
133 -- 2023-06-07T12:17:51.000000000
134 -- 2023-06-07T13:23:18.000000000
135 -- 2023-06-07T14:28:46.000000000
136 -- 2023-06-07T15:34:13.000000000
137 -- 2023-06-07T16:39:40.000000000
138 -- 2023-06-07T17:45:07.000000000
139 -- 2023-06-07T18:50:34.000000000
140 -- 2023-06-07T19:56:02.000000000
141 -- 2023-06-07T21:01:30.000000000
142 -- 2023-06-07T22:06:56.000000000
143 -- 2023-06-07T23:12:23.000000000
144 -- 2023-06-08T00:08:30.000000000
145 -- 2023-06-08T01:13:58.000000000
146 -- 2023-06-08T02:19:25.000000000
147 -- 2023-06-08T03:24:52.000000000
148 -- 2023-06-08T04:30:19.000000000
149 -- 2023-06-08T05:35:47.000000000
150 -- 2023-06-08T06:41:14.000000000
151 -- 2023-06-08T07:46:41.000000000
152 -- 2023-06-08T08:52:08.000000000
153 -- 2023-06-08T09:57:35.000000000
154 -- 2023-06-08T11:03:02.000000000
155 -- 2023-06-08T12:08:30.000000000
156 -- 2023-06-08T13:13:57.000000000
157 -- 2023-06-08T14:19:25.000000000
158 -- 2023-06-08T15:24:52.000000000
159 -- 2023-06-08T16:30:20.000000000
160 -- 2023-06-08T17:35:47.000000000
161 -- 2023-06-08T18:41:14.000000000
162 -- 2023-06-08T19:46:42.000000000
163 -- 2023-06-08T20:52:10.000000000
164 -- 2023-06-08T21:57:36.000000000
165 -- 2023-06-08T23:03:04.000000000
166 -- 2023-06-09T00:27:13.000000000
167 -- 2023-06-09T01:32:40.000000000
168 -- 2023-06-09T02:38:07.000000000
169 -- 2023-06-09T03:43:34.000000000
170 -- 2023-06-09T04:49:01.000000000
171 -- 2023-06-09T05:54:29.000000000
172 -- 2023-06-09T06:59:56.000000000
173 -- 2023-06-09T08:05:23.000000000
174 -- 2023-06-09T09:10:50.000000000
175 -- 2023-06-09T10:16:18.000000000
176 -- 2023-06-09T11:21:45.000000000
177 -- 2023-06-09T12:27:13.000000000
178 -- 2023-06-09T13:32:40.000000000
179 -- 2023-06-09T14:38:07.000000000
180 -- 2023-06-09T15:43:34.000000000
181 -- 2023-06-09T16:49:02.000000000
182 -- 2023-06-09T17:54:29.000000000
183 -- 2023-06-09T18:59:57.000000000
184 -- 2023-06-09T20:05:24.000000000
185 -- 2023-06-09T21:10:52.000000000
186 -- 2023-06-09T22:16:19.000000000
187 -- 2023-06-09T23:21:46.000000000
188 -- 2023-06-10T00:45:54.000000000
189 -- 2023-06-10T01:51:22.000000000
190 -- 2023-06-10T02:56:49.000000000
191 -- 2023-06-10T04:02:16.000000000
192 -- 2023-06-10T05:07:44.000000000
193 -- 2023-06-10T06:13:11.000000000
194 -- 2023-06-10T07:18:38.000000000
195 -- 2023-06-10T08:24:05.000000000
196 -- 2023-06-10T09:29:33.000000000
197 -- 2023-06-10T10:35:00.000000000
198 -- 2023-06-10T11:40:28.000000000
199 -- 2023-06-10T12:45:55.000000000
200 -- 2023-06-10T13:51:23.000000000
201 -- 2023-06-10T14:56:50.000000000
202 -- 2023-06-10T16:02:17.000000000
203 -- 2023-06-10T17:07:45.000000000
204 -- 2023-06-10T18:13:12.000000000
205 -- 2023-06-10T19:18:40.000000000
206 -- 2023-06-10T20:24:08.000000000
207 -- 2023-06-10T21:29:36.000000000
208 -- 2023-06-10T22:35:02.000000000
209 -- 2023-06-10T23:40:30.000000000
210 -- 2023-06-11T01:04:36.000000000
211 -- 2023-06-11T02:10:04.000000000
212 -- 2023-06-11T03:15:31.000000000
213 -- 2023-06-11T04:20:59.000000000
214 -- 2023-06-11T05:26:27.000000000
215 -- 2023-06-11T06:31:54.000000000
216 -- 2023-06-11T07:37:21.000000000
217 -- 2023-06-11T08:42:48.000000000
218 -- 2023-06-11T09:48:16.000000000
219 -- 2023-06-11T10:53:43.000000000
220 -- 2023-06-11T11:59:11.000000000
221 -- 2023-06-11T13:04:38.000000000
222 -- 2023-06-11T14:10:05.000000000
223 -- 2023-06-11T15:15:33.000000000
224 -- 2023-06-11T16:21:00.000000000
225 -- 2023-06-11T17:26:28.000000000
226 -- 2023-06-11T18:31:55.000000000
227 -- 2023-06-11T19:37:22.000000000
228 -- 2023-06-11T20:42:49.000000000
229 -- 2023-06-11T21:48:18.000000000
230 -- 2023-06-11T22:53:44.000000000
231 -- 2023-06-12T00:55:15.000000000
232 -- 2023-06-12T02:00:43.000000000
233 -- 2023-06-12T03:06:11.000000000
234 -- 2023-06-12T04:11:38.000000000
235 -- 2023-06-12T05:17:05.000000000
236 -- 2023-06-12T06:22:32.000000000
237 -- 2023-06-12T07:27:59.000000000
238 -- 2023-06-12T08:33:26.000000000
239 -- 2023-06-12T09:38:54.000000000
240 -- 2023-06-12T10:44:22.000000000
241 -- 2023-06-12T11:49:49.000000000
242 -- 2023-06-12T12:55:16.000000000
243 -- 2023-06-12T14:00:44.000000000
244 -- 2023-06-12T15:06:11.000000000
245 -- 2023-06-12T16:11:39.000000000
246 -- 2023-06-12T17:17:06.000000000
247 -- 2023-06-12T18:22:33.000000000
248 -- 2023-06-12T19:28:01.000000000
249 -- 2023-06-12T20:33:28.000000000
250 -- 2023-06-12T21:38:56.000000000
251 -- 2023-06-12T22:44:23.000000000
252 -- 2023-06-12T23:49:50.000000000
253 -- 2023-06-13T00:36:34.000000000
254 -- 2023-06-13T01:42:01.000000000
255 -- 2023-06-13T02:47:28.000000000
256 -- 2023-06-13T03:52:56.000000000
257 -- 2023-06-13T04:58:23.000000000
258 -- 2023-06-13T06:03:50.000000000
259 -- 2023-06-13T07:09:17.000000000
260 -- 2023-06-13T08:14:44.000000000
261 -- 2023-06-13T09:20:11.000000000
262 -- 2023-06-13T10:25:39.000000000
263 -- 2023-06-13T11:31:06.000000000
264 -- 2023-06-13T12:36:33.000000000
265 -- 2023-06-13T13:42:00.000000000
266 -- 2023-06-13T14:47:28.000000000
267 -- 2023-06-13T15:52:55.000000000
268 -- 2023-06-13T16:58:23.000000000
269 -- 2023-06-13T18:03:50.000000000
270 -- 2023-06-13T19:09:17.000000000
271 -- 2023-06-13T20:14:44.000000000
272 -- 2023-06-13T21:20:13.000000000
273 -- 2023-06-13T22:25:39.000000000
274 -- 2023-06-13T23:31:06.000000000
275 -- 2023-06-14T00:17:52.000000000
276 -- 2023-06-14T01:23:19.000000000
277 -- 2023-06-14T02:28:46.000000000
278 -- 2023-06-14T03:34:14.000000000
279 -- 2023-06-14T04:39:41.000000000
280 -- 2023-06-14T05:45:08.000000000
281 -- 2023-06-14T07:56:03.000000000
282 -- 2023-06-14T09:01:30.000000000
283 -- 2023-06-14T10:06:57.000000000
284 -- 2023-06-14T11:12:24.000000000
285 -- 2023-06-14T12:17:51.000000000
286 -- 2023-06-14T13:23:19.000000000
287 -- 2023-06-14T14:28:46.000000000
288 -- 2023-06-14T15:34:13.000000000
289 -- 2023-06-14T16:39:40.000000000
290 -- 2023-06-14T17:44:41.000000000
291 -- 2023-06-14T17:45:07.000000000
292 -- 2023-06-14T18:50:34.000000000
293 -- 2023-06-14T19:56:02.000000000
294 -- 2023-06-14T21:01:29.000000000
295 -- 2023-06-14T22:06:57.000000000
296 -- 2023-06-14T23:12:24.000000000
297 -- 2023-06-15T00:08:30.000000000
298 -- 2023-06-15T01:13:57.000000000
299 -- 2023-06-15T02:19:24.000000000
300 -- 2023-06-15T03:24:52.000000000
301 -- 2023-06-15T04:30:19.000000000
302 -- 2023-06-15T05:35:46.000000000
303 -- 2023-06-15T06:41:14.000000000
304 -- 2023-06-15T07:46:41.000000000
305 -- 2023-06-15T08:52:08.000000000
306 -- 2023-06-15T09:57:35.000000000
307 -- 2023-06-15T11:03:03.000000000
308 -- 2023-06-15T12:08:30.000000000
309 -- 2023-06-15T13:13:57.000000000
310 -- 2023-06-15T14:19:25.000000000
311 -- 2023-06-15T15:24:52.000000000
312 -- 2023-06-15T16:30:20.000000000
313 -- 2023-06-15T17:35:47.000000000
314 -- 2023-06-15T18:41:14.000000000
315 -- 2023-06-15T19:46:41.000000000
316 -- 2023-06-15T20:52:09.000000000
317 -- 2023-06-15T21:57:36.000000000
318 -- 2023-06-15T23:03:03.000000000
319 -- 2023-06-16T00:27:13.000000000
320 -- 2023-06-16T01:32:40.000000000
321 -- 2023-06-16T02:38:08.000000000
322 -- 2023-06-16T03:43:35.000000000
323 -- 2023-06-16T04:49:02.000000000
324 -- 2023-06-16T05:54:29.000000000
325 -- 2023-06-16T06:59:56.000000000
326 -- 2023-06-16T08:05:23.000000000
327 -- 2023-06-16T09:10:50.000000000
328 -- 2023-06-16T10:16:18.000000000
329 -- 2023-06-16T11:21:45.000000000
330 -- 2023-06-16T12:27:13.000000000
331 -- 2023-06-16T13:32:40.000000000
332 -- 2023-06-16T14:38:07.000000000
333 -- 2023-06-16T15:43:34.000000000
334 -- 2023-06-16T16:49:01.000000000
335 -- 2023-06-16T17:54:29.000000000
336 -- 2023-06-16T18:59:56.000000000
337 -- 2023-06-16T20:05:23.000000000
338 -- 2023-06-16T21:10:51.000000000
339 -- 2023-06-16T22:16:18.000000000
340 -- 2023-06-16T23:21:45.000000000# Choose the timestep here.
timestep = 53Get the data arrays for the selected timestep
geo_mask = get_geo_mask(mfds['Longitude'][timestep, :, :], mfds['Latitude'][timestep, :, :]).compute().values
lon_values = mfds['Longitude'][timestep, :, :].where(geo_mask).values
lat_values = mfds['Latitude'][timestep, :, :].where(geo_mask).values
var_values = mfds[data_variable][timestep, :, :].where(geo_mask).values
timestamp = datetime.strptime(
np.datetime_as_string(mfds['time'][timestep].values, unit='ms'),
"%Y-%m-%dT%H:%M:%S.%f"
)# Get value range for the single timestep
values_min = 0 # Alternatively, one could use `np.nanmin(var_values)`.
values_max = 10
print(f"{values_min} to {values_max}")0 to 10# country boundaries
country_bodr = cfeature.NaturalEarthFeature(category='cultural',
name='admin_0_boundary_lines_land', scale="110m", facecolor='none', edgecolor='k')
# province boundaries
provinc_bodr = cfeature.NaturalEarthFeature(category='cultural',
name='admin_1_states_provinces_lines', scale="110m", facecolor='none', edgecolor='k')my_projection = ccrs.PlateCarree()
fig, ax = plt.subplots(figsize=(12, 9), subplot_kw={"projection": my_projection}) #, "extent": extent})
vmin, vmax = values_min, values_max
levels = 50
level_boundaries = np.linspace(vmin, vmax, levels + 1)
_contourf = ax.contourf(lon_values, lat_values, var_values,
cmap=my_turbo, transform=ccrs.PlateCarree(),
levels=level_boundaries,
vmin=vmin, vmax=vmax, extend='both')
ax.add_feature(cfeature.STATES, linestyle='--', linewidth=1, edgecolor="w")
cb_handle = plt.colorbar(
_contourf,
orientation="horizontal",
ticks=range(int(np.floor(vmin)), int(np.ceil(vmax+1)), 1),
boundaries=level_boundaries,
values=(level_boundaries[:-1] + level_boundaries[1:]) / 2,
ax=ax
)
cb_handle.ax.set_title(data_variable, fontsize=18)
ax.set_title("%s" % (timestamp.strftime("%Y%m%d %H:%M:%S")), fontsize=18)
gl_handle = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=['left', 'bottom'],
linewidth=0.8, color='gray', alpha=0.5, linestyle=':')
# We change the fontsize tick labels
ax.tick_params(axis='both', which='major', labelsize=16)
ax.tick_params(axis='both', which='minor', labelsize=16)
cb_handle.ax.tick_params(axis='both', which='minor', labelsize=16)
gl_handle.xlabel_style, gl_handle.ylabel_style = {'fontsize': 16}, {'fontsize': 16}
# plt.savefig("UVAI-%s" % timestamp.strftime("%Y%m%d%H%M%S"), bbox_inches="tight")
plt.show()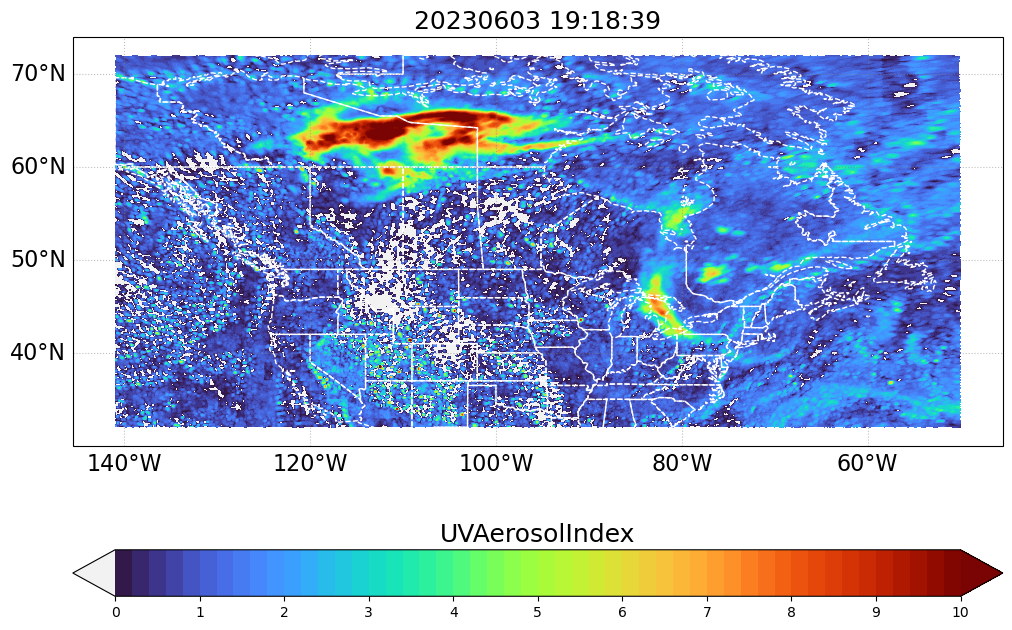
Remake the same map but with masked values
Here we specify the UVAI threshold for which we will cut off the values.
mx = ma.masked_array(var_values, mask=(var_values < 4))my_projection = ccrs.PlateCarree()
fig, ax = plt.subplots(figsize=(12, 9), subplot_kw={"projection": my_projection})
vmin, vmax = values_min, values_max
levels = 50
level_boundaries = np.linspace(vmin, vmax, levels + 1)
_contourf = ax.contourf(lon_values, lat_values, mx,
cmap=my_turbo, transform=ccrs.PlateCarree(),
levels=level_boundaries,
vmin=vmin, vmax=vmax, extend='both')
ax.add_feature(cfeature.STATES, linestyle='--', linewidth=0.8, edgecolor="k")
cb_handle = plt.colorbar(
_contourf,
orientation="horizontal",
ticks=range(int(np.floor(vmin)), int(np.ceil(vmax+1)), 1),
boundaries=level_boundaries,
values=(level_boundaries[:-1] + level_boundaries[1:]) / 2,
ax=ax
)
cb_handle.ax.set_title(data_variable, fontsize=18)
ax.set_title("%s" % (timestamp.strftime("%Y%m%d %H:%M:%S")), fontsize=18)
gl_handle = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=['left', 'bottom'],
linewidth=0.8, color='gray', alpha=0.5, linestyle=':')
# We change the fontsize tick labels
ax.tick_params(axis='both', which='major', labelsize=16)
ax.tick_params(axis='both', which='minor', labelsize=16)
cb_handle.ax.tick_params(axis='both', which='minor', labelsize=16)
gl_handle.xlabel_style, gl_handle.ylabel_style = {'fontsize': 16}, {'fontsize': 16}
plt.show()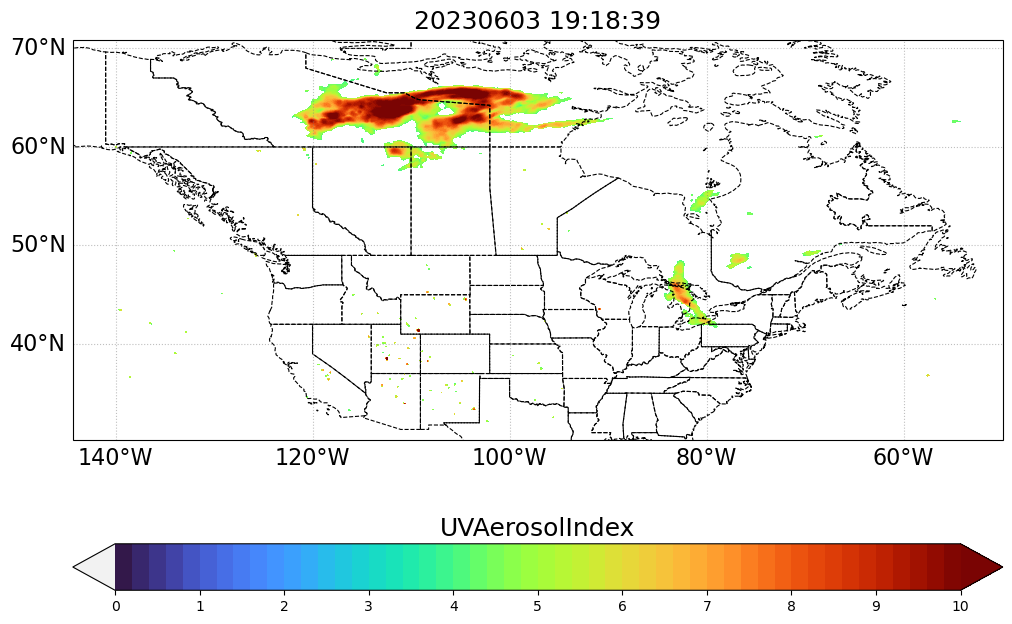
5. Make multi-panel figure of three timesteps
my_projection = ccrs.PlateCarree()
fig, axs = plt.subplots(figsize=(10, 16), nrows=3, sharex=True, sharey=True,
subplot_kw={"projection": my_projection}) #, "extent": extent})
vmin, vmax = values_min, values_max
levels = 50
level_boundaries = np.linspace(vmin, vmax, levels + 1)
timesteps = [93, 116, 139]
for i, (ax, ts) in enumerate(zip(axs, timesteps)):
print(f"i - {ts}")
lon_values, lat_values, var_values, timestamp_str = get_data_arrays(ts)
_contourf = ax.contourf(lon_values, lat_values, var_values,
cmap=my_turbo, transform=ccrs.PlateCarree(),
levels=level_boundaries,
vmin=vmin, vmax=vmax, extend='both')
ax.add_feature(cfeature.STATES, linestyle='--', linewidth=1, edgecolor="w")
ax.set_title(
"%s" % (timestamp_str.strftime("%Y%m%d %H:%M:%S")), fontsize=18
)
if i < 2:
grid_labels_locations = ['left']
else:
grid_labels_locations = ['left', 'bottom']
gl_handle = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=grid_labels_locations,
linewidth=0.8, color='gray', alpha=0.5, linestyle=':')
# We change the fontsize tick labels
ax.tick_params(axis='both', which='major', labelsize=16)
ax.tick_params(axis='both', which='minor', labelsize=16)
gl_handle.xlabel_style, gl_handle.ylabel_style = {'fontsize': 16}, {'fontsize': 16}
cb_handle = fig.colorbar(
_contourf,
orientation="horizontal",
ticks=range(int(np.floor(vmin)), int(np.ceil(vmax+1)), 1),
boundaries=level_boundaries,
values=(level_boundaries[:-1] + level_boundaries[1:]) / 2,
ax=axs,
pad=0.07
)
cb_handle.ax.set_title(data_variable, fontsize=18)
cb_handle.ax.tick_params(axis='both', which='minor', labelsize=16)
# plt.savefig("UVAI-multiple.png", bbox_inches="tight", dpi=250)
plt.show()i - 93
i - 116
i - 139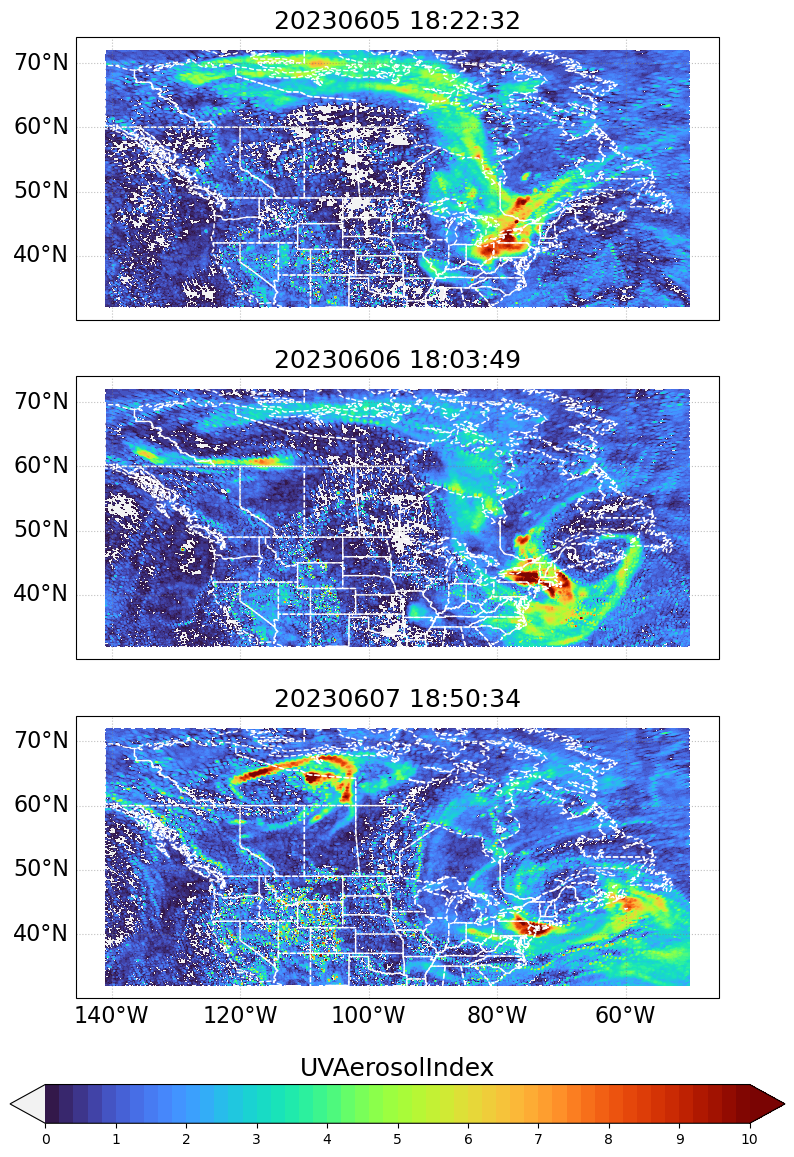
# 6. Make a timeseries figure
Find point to represent Manhattan
times = [datetime.strptime(np.datetime_as_string(t.values, unit='ms'), "%Y-%m-%dT%H:%M:%S.%f")
for t in mfds['time']]
n_time = len(mfds.time)def get_manhattan_value(a_timestep: int, query_point_lon_lat: tuple):
lon_values, lat_values, var_values, timestamp = get_data_arrays(a_timestep)
lon_flat, lat_flat = lon_values.flatten(), lat_values.flatten()
new2d = np.zeros((len(lon_flat), 2))
for idx, (a, b) in enumerate(zip(lon_flat, lat_flat)):
if not np.isnan(a):
new2d[idx, :] = [a, b]
nearestpt_distance, nearestpt_index = spatial.KDTree(new2d).query(query_point_lon_lat)
nearestpt_coords = new2d[nearestpt_index, :]
return nearestpt_distance, nearestpt_index, nearestpt_coords!! Warning !! This next cell can take a long time (e.g., +20 minutes). It needs to be run once, but then should be commented-out.
# # 40.7831° N, 73.9712° W
# pt_manhattan = [-73.9712, 40.7831]
# nearestpt_distances = []
# nearestpt_indices = []
# nearestpt_coords = []
# for i in tqdm(range(n_time)):
# nearest_distance, nearest_index, nearest_coords = get_manhattan_value(i, pt_manhattan)
# nearestpt_distances.append(nearest_distance)
# nearestpt_indices.append(nearest_index)
# nearestpt_coords.append(nearest_coords)
# # Now save the results to JSON files so you don't have to run this again
# with open("nearestpt_distances.json", "w") as fp:
# json.dump(nearestpt_distances, fp)
# with open("nearestpt_indices.json", "w") as fp:
# json.dump([int(i) for i in nearestpt_indices], fp)
# with open("nearestpt_coords.json", "w") as fp:
# json.dump([[i[0], i[1]] for i in nearestpt_coords], fp)Load the values stored during execution of the previous cell.
with open("nearestpt_distances.json", "r") as fp:
nearestpt_distances = json.load(fp)
with open("nearestpt_indices.json", "r") as fp:
nearestpt_indices = json.load(fp)
with open("nearestpt_coords.json", "r") as fp:
nearestpt_coords = json.load(fp)In this next cell, we extract the actual data values at the indices belonging to the point closest to Manhattan. This cell takes just a few minutes.
timeseries_values = []
for i in tqdm(range(n_time)):
_, _, var_values, timestamp = get_data_arrays(i)
nearestpt_var_value = var_values.flatten()[nearestpt_indices[i]]
timeseries_values.append(nearestpt_var_value)Generate figure for DSCOVR EPIC
fig, ax = plt.subplots(figsize=(12, 9))
ax.plot(times, timeseries_values, ":o", markersize=7, linewidth=2)
ax.set_ylabel(f"{data_variable} @Manhattan, NY\n(DSCOVR_EPIC_L2_AER)", fontsize=18)
# ax.set_ylim((-10, 10))
# We change the fontsize tick labels
ax.tick_params(axis='both', which='major', labelsize=16)
ax.tick_params(axis='both', which='minor', labelsize=16)
plt.xticks(rotation=45)
# plt.savefig("dscovr_epic_uvai_timeseries_20230601-20230617.png",
# dpi=200, bbox_inches="tight")
plt.show()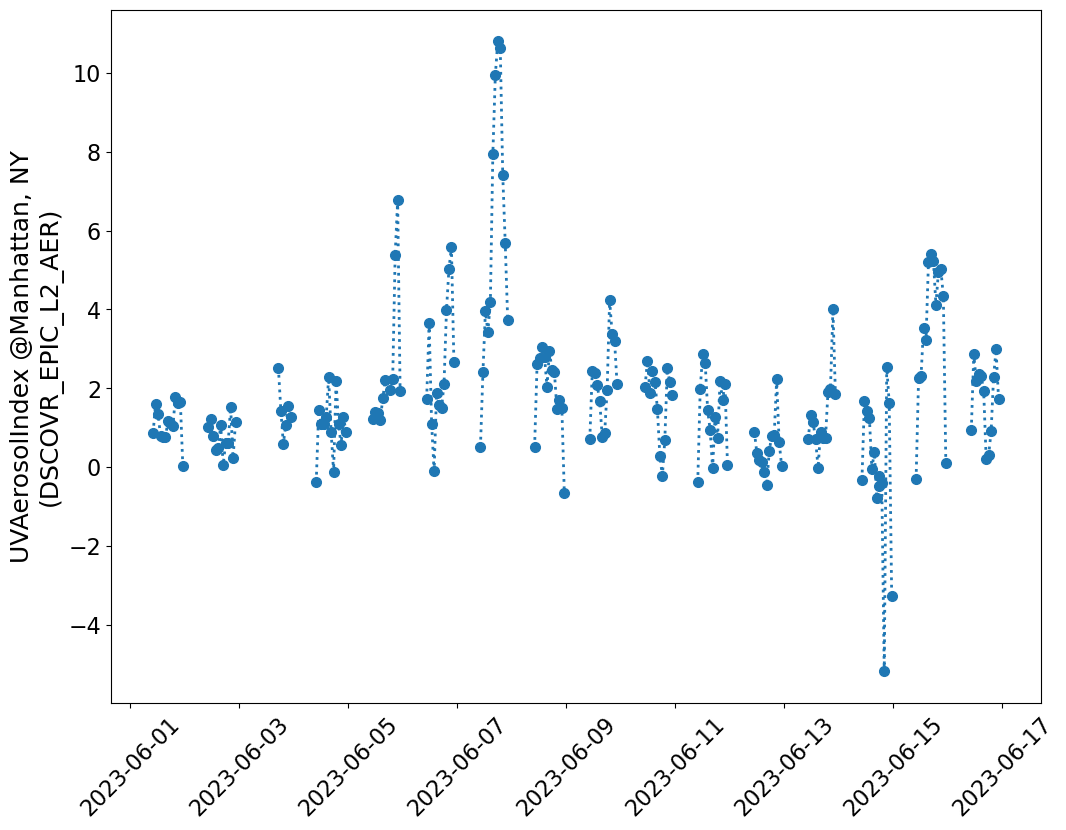
Use RSIG to get EPA Air Quality data from the same time period
rsigapi = pyrsig.RsigApi(
bdate='2023-06-01 00',
edate='2023-06-17 23:59:59',
bbox=(-74.05, 40.68, -73.8, 40.90) #Manhattan
)
aqsno2df = rsigapi.to_dataframe('aqs.pm25', parse_dates=True, unit_keys=False)
aqsno2s = aqsno2df.groupby(['time']).median(numeric_only=True)['pm25']Make combined figure
fig, axs = plt.subplots(figsize=(10, 12), nrows=2, ncols=1, sharex=True)
# Plot
# tax = ax.twinx()
axn = 0
axs[axn].plot(
aqsno2s.index.values, aqsno2s.values, "-o", markersize=3, color='gray'
)
axs[axn].set_ylabel(f"Manhattan, NY\n(AQS pm25)", fontsize=18, color='k')
axn = 1
axs[axn].plot(
times, timeseries_values, "-o", markersize=3, color="gray"
)
axs[axn].set_ylabel(f"{data_variable} @Manhattan, NY\n(DSCOVR_EPIC_L2_AER)", fontsize=18, color="black")
# Change the aesthetics
for a in axs:
a.axvspan(datetime(2023, 6, 6, 0, 0, 0),
datetime(2023, 6, 10, 0, 0, 0),
alpha=0.3, color='pink')
a.axhline(0, linestyle=":", alpha=0.3, color="gray")
a.tick_params(axis='both', which='major', labelsize=16)
a.tick_params(axis='both', which='minor', labelsize=16)
a.grid(visible=True, which='major', axis='both', color="lightgray", linestyle=":")
a.tick_params(color='gray', labelcolor='black')
for spine in a.spines.values():
spine.set_edgecolor('gray')
plt.xticks(rotation=45)
plt.tight_layout()
# plt.savefig("dscovr_epic_uvai_with_AQS_timeseries_20230601-20230617.png",
# dpi=200, bbox_inches="tight")
plt.show()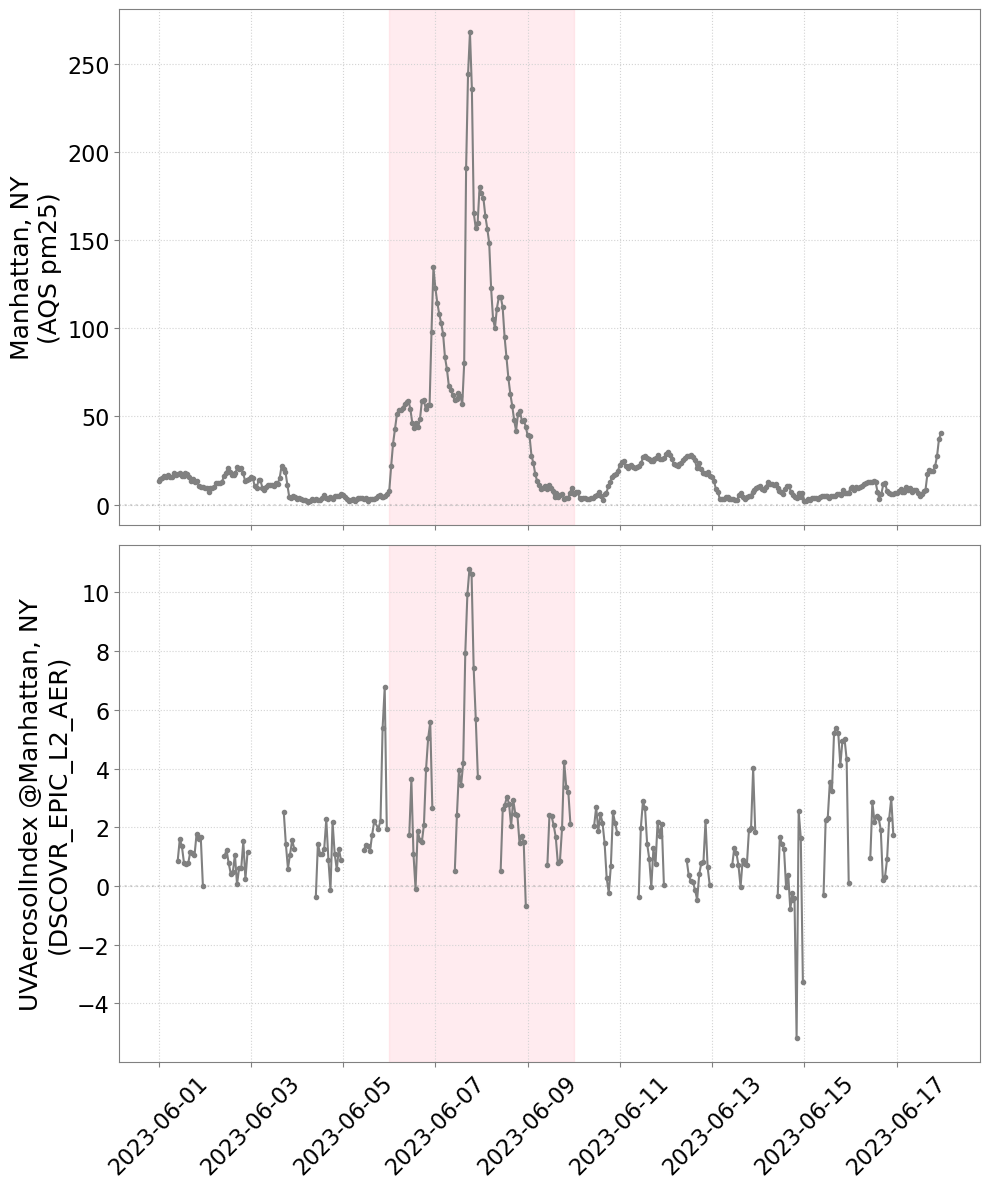
7. Create an animation
# Get overall min and max
overall_geo_mask = get_geo_mask(mfds['Longitude'], mfds['Latitude']).compute().values
overall_var_values = mfds[data_variable].where(overall_geo_mask).values
# overall_values_min = np.nanmin(overall_var_values)
# overall_values_max = np.nanmax(overall_var_values[overall_var_values != np.inf])
overall_values_min = 0
overall_values_max = 10
print(f"{overall_values_min} to {overall_values_max}")0 to 10fig, ax = plt.subplots(figsize=(12, 9), subplot_kw={"projection": ccrs.PlateCarree()})
vmin, vmax = overall_values_min, overall_values_max
levels = 50
level_boundaries = np.linspace(vmin, vmax, levels + 1)
# first image on screen
lon_values, lat_values, var_values, timestamp = get_data_arrays(0)
_contourf = ax.contourf(lon_values, lat_values, var_values,
cmap=my_turbo, transform=ccrs.PlateCarree(),
levels=level_boundaries, extend='both',
vmin=vmin, vmax=vmax)
plt.colorbar(
_contourf,
ticks=range(int(np.floor(vmin)), int(np.ceil(vmax+1)), 1),
boundaries=level_boundaries,
values=(level_boundaries[:-1] + level_boundaries[1:]) / 2,
ax=ax
)
# ax.add_feature(cfeature.STATES, edgecolor='black', linewidth=1)
ax.add_feature(cfeature.STATES, linestyle='--', linewidth=0.8, edgecolor="k")
ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=['left', 'bottom'],
linewidth=1, color='gray', alpha=0.5, linestyle=':')
# animation function
def animate(i):
global _contourf
lon_values, lat_values, var_values, timestamp = get_data_arrays(i)
for c in _contourf.collections:
c.remove() # removes only the contours, leaves the rest intact
_contourf = ax.contourf(lon_values, lat_values, var_values,
cmap=my_turbo, transform=ccrs.PlateCarree(),
levels=level_boundaries, extend='both',
vmin=vmin, vmax=vmax)
ax.set_title("%s\n\nFrame %s - %s" %
(data_variable, str(i), timestamp.strftime("%Y %m %d %H:%M:%S")))
return _contourf
n_frames = 53
ani = FuncAnimation(fig, animate, repeat=False, frames=n_frames, )
# ani.save('UVAI_latest.gif', writer='imagemagick', fps=2, progress_callback=lambda i, n: print(f"{i:03}/{n_frames}"))/var/folders/m6/n4zwmdm91l52f9d9_sj49zy00000gp/T/ipykernel_56884/565891392.py:33: MatplotlibDeprecationWarning: The collections attribute was deprecated in Matplotlib 3.8 and will be removed two minor releases later.
for c in _contourf.collections:
END of Notebook.
