import netCDF4
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import matplotlib.axes
import cartopy
import cartopy.crs as ccrs
import cartopy.feature as cfeat
from optparse import OptionParser
from netCDF4 import Dataset
from IPython.display import HTMLPlotting MAIA netCDF-4 PM Level 2 Data Variables
Acknowledgements
The research was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with the National Aeronautics and Space Administration (80NM0018D0004).
© 2021 California Institute of Technology. Government sponsorship acknowledged.
Getting Started
In this notebook, we will access a local file of simulated MAIA Level 2 PM data. These simulated data are provided courtesy of the MAIA Project in netCDF-4 format.
We will access the data file from local storage, load data variables into memory, and plot the data on a 2-D regional map over the Primary Target Area (PTA) of Los Angeles (USA).
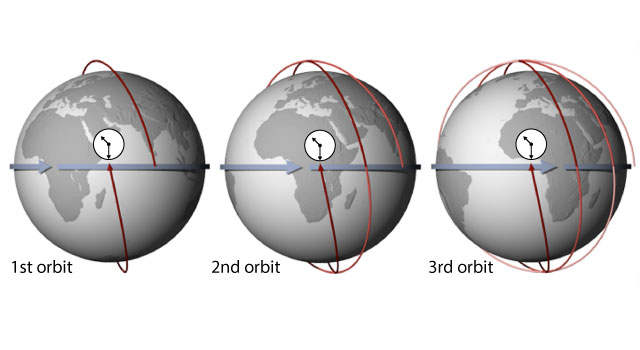
MAIA Orbital Targetting Simulation
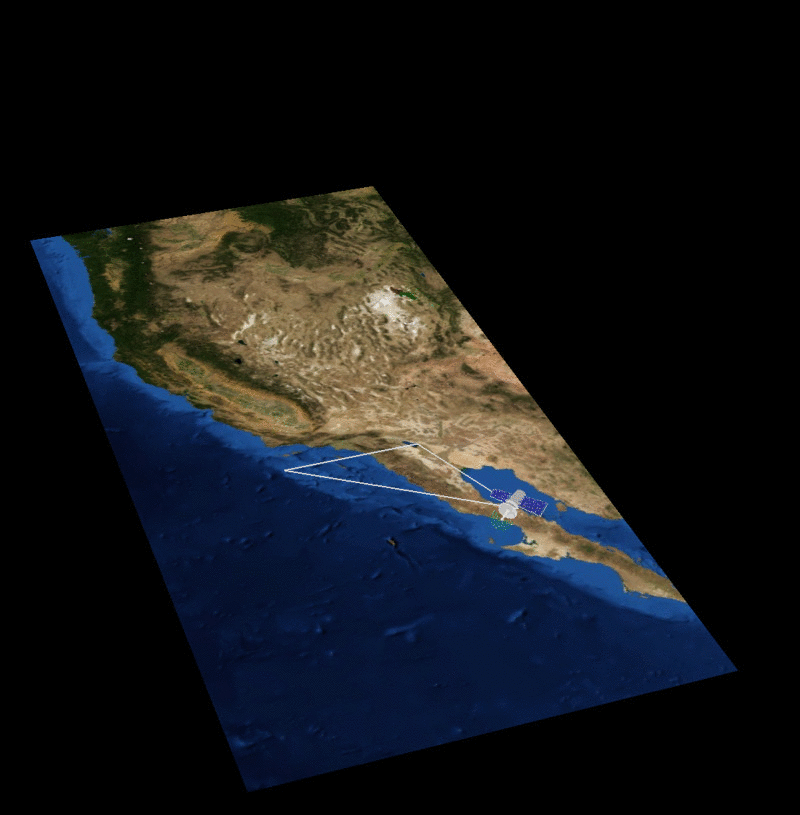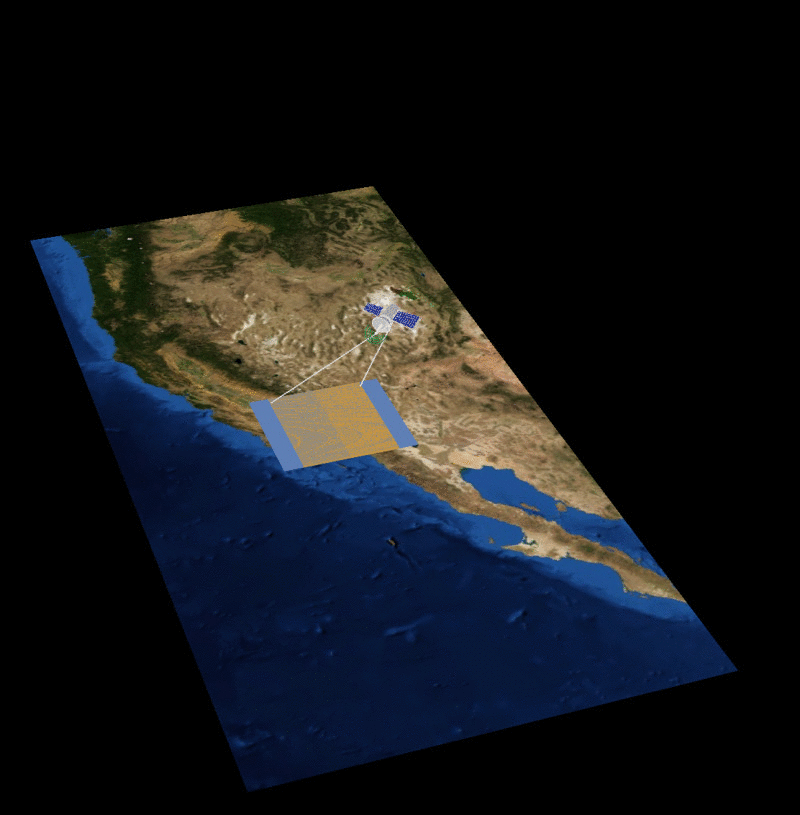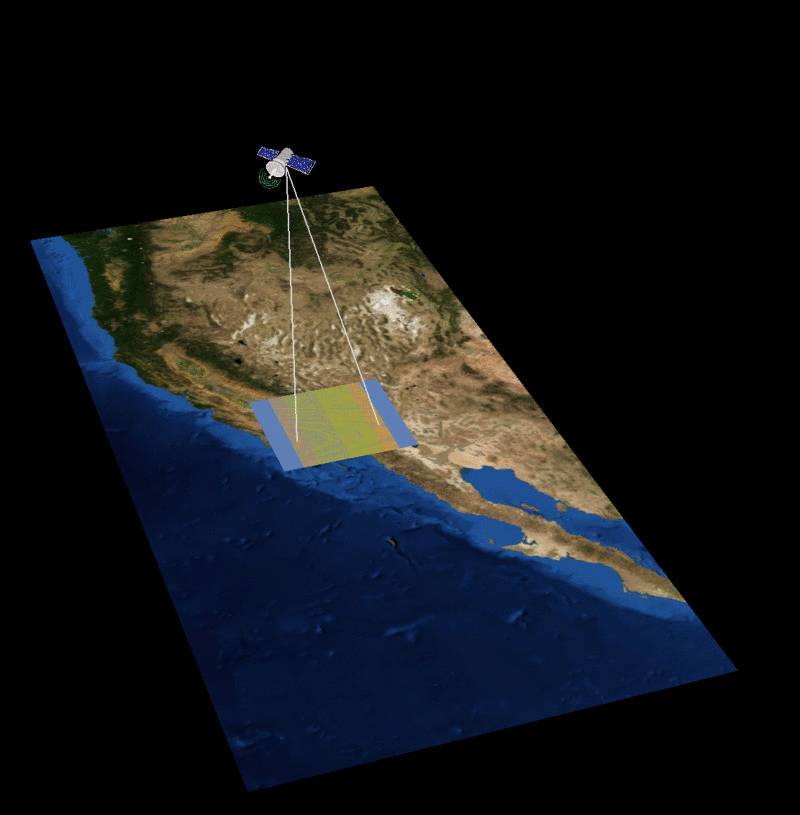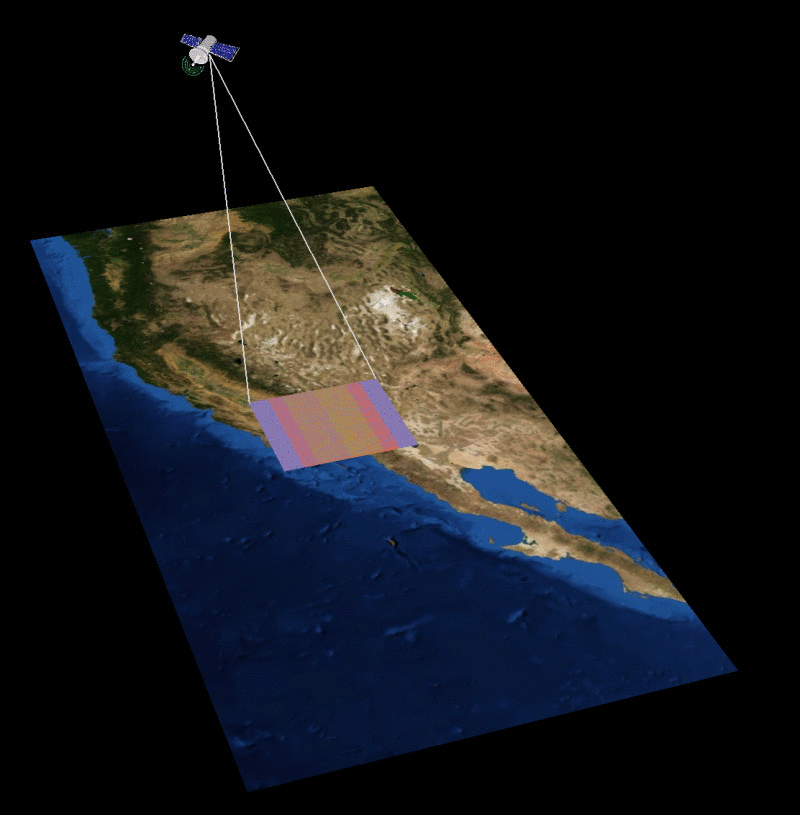
MAIA Sequential Orbit Time
Requirements
Python 3
Most of these imports are from the Python standard library. However, you will need to install these packages into your Python 3 environment if you have not already done so:
- netCDF4
- numpy
- matplotlib
- mpl_toolkits
- cartopy
- optparse
- IPython
Read the netCDF file into memory
pm_file = "MAIA_L2_PM_PM-10-total_20190102T000000Z_XX_XXX_RXX_USA-LosAngeles_FXX_VXXpXXpXXpXX.nc"
print ( pm_file )
# -----------------------------
# Open netCDF4 file for reading
# -----------------------------
try:
nc_file = Dataset( pm_file, 'r' )
except IOError:
print ('not a valid netCDF file')MAIA_L2_PM_PM-10-total_20190102T000000Z_XX_XXX_RXX_USA-LosAngeles_FXX_VXXpXXpXXpXX.ncOpen with the netCDF file, starting with reading in the Global Attributes.
def readGlobalAttrs( nc_file ):
attr_list = []
global_attr = []
for attr_name in nc_file.ncattrs():
# print ( attr_name, '=', getattr(nc_file, attr_name) )
attr_list.append( attr_name )
atts = getattr( nc_file, attr_name )
global_attr.append( atts )
return attr_list, global_attr Print the netCDF Global Attributes
print ('Global attributes:')
[attr_list, global_attr] = readGlobalAttrs( nc_file )
natts = len(attr_list)
for n in range(0, natts):
print (attr_list[n]," = ",global_attr[n])Global attributes:
title = GRM Level 2 Stage 2 PM Product
source = GRM Level 2 Stage 2 Daily PM Product is the result of application of GRM Level 2 Stage 1 regression coefficients to L2 aerosol retrievals and other ancillary data.
history = <date>: Initial production using software version <version tag>, built <build date>, by <user id>. See also Software_version_information and Input_files.
institution = MAIA products are generated by the MAIA Science Team, using processing and storage facilities of the NASA Langley Research Center Atmospheric Science Data Center (ASDC).
data_start_time = YYYY-MM-DDTHH:MM:SS.sssZ
data_end_time = YYYY-MM-DDTHH:MM:SS.sssZ
collection_number = CXX
local_granule_id = MAIA_[ProductLevel]_[ProductName]_[DateTimeStamp]_[ProcessingStrategy]_[QualityDesignator]_[ReprocessingVersion]_[TargetName]_[FormatVersion]_[DataVersion].nc
target_name =
input_files = A,B,C,...,X,Y,Z
coremetadata = <gmi:MI_Metadata xmlns:eos="http://earthdata.nasa.gov/schema/eos"
xmlns:gco="http://www.isotc211.org/2005/gco" xmlns:gmd="http://www.isotc211.org/2005/gmd"
xmlns:gmi="http://www.isotc211.org/2005/gmi" xmlns:gml="http://www.opengis.net/gml/3.2"
xmlns:gmx="http://www.isotc211.org/2005/gmx" xmlns:gsr="http://www.isotc211.org/2005/gsr"
xmlns:gss="http://www.isotc211.org/2005/gss" xmlns:gts="http://www.isotc211.org/2005/gts"
xmlns:srv="http://www.isotc211.org/2005/srv" xmlns:xlink="http://www.w3.org/1999/xlink"
xmlns:xs="http://www.w3.org/2001/XMLSchema" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://earthdata.nasa.gov/schema/eos https://cdn.earthdata.nasa.gov/iso/eos/1.0/eos.xsd
http://www.isotc211.org/2005/gco https://cdn.earthdata.nasa.gov/iso/gco/1.0/gco.xsd
http://www.isotc211.org/2005/gmd https://cdn.earthdata.nasa.gov/iso/gmd/1.0/gmd.xsd
http://www.isotc211.org/2005/gmi https://cdn.earthdata.nasa.gov/iso/gmi/1.0/gmi.xsd
http://www.opengis.net/gml/3.2 https://cdn.earthdata.nasa.gov/iso/gml/1.0/gml.xsd
http://www.isotc211.org/2005/gmx https://cdn.earthdata.nasa.gov/iso/gmx/1.0/gmx.xsd
http://www.isotc211.org/2005/gsr https://cdn.earthdata.nasa.gov/iso/gsr/1.0/gsr.xsd
http://www.isotc211.org/2005/gss https://cdn.earthdata.nasa.gov/iso/gss/1.0/gss.xsd
http://www.isotc211.org/2005/gts https://cdn.earthdata.nasa.gov/iso/gts/1.0/gts.xsd
http://www.isotc211.org/2005/srv https://cdn.earthdata.nasa.gov/iso/srv/1.0/srv.xsd">
<!-- ..................................................................................
ISO-MENDS Template for MAIA L1B2 products
2019-10-28 Version 1.2
-->
<!-- Change log .........................................................................
2020-04-08 Template updated per entries made by MAIA science team in metadata table at https://wiki.earthdata.nasa.gov/display/maia/L2+PM+Granule+Metadata%3A+ISO+MENDS
-->
<!-- The granule file name. -->
<gmd:fileIdentifier>
<gco:CharacterString>MAIA_L2_PM_PM-10-total_20190102T000000Z_XX_XXX_RXX_USA-LosAngeles_FXX_VXXpXXpXXpXX.nc</gco:CharacterString>
</gmd:fileIdentifier>
<!-- The language used as the content of this metadata record. -->
<gmd:language>
<gco:CharacterString>eng</gco:CharacterString>
</gmd:language>
<!-- The character set used in this metadata record.-->
<gmd:characterSet>
<gmd:MD_CharacterSetCode codeList="http://www.ngdc.noaa.gov/metadata/published/xsd/schema/resources/Codelist/gmxCodelists.xml#MD_CharacterSetCode" codeListValue="utf8">utf8</gmd:MD_CharacterSetCode>
</gmd:characterSet>
<!-- What is represented by this metadata record - currently series means collection and dataset means granule.-->
<gmd:hierarchyLevel>
<gmd:MD_ScopeCode codeList="http://www.ngdc.noaa.gov/metadata/published/xsd/schema/resources/Codelist/gmxCodelists.xml#MD_ScopeCode" codeListValue="dataset">dataset</gmd:MD_ScopeCode>
</gmd:hierarchyLevel>
<!-- There is no contact information that is read or generated by the CMR for granules -->
<gmd:contact gco:nilReason="missing"/>
<!-- This section just documents the ISO schema and version used for this record. -->
<gmd:metadataStandardName>
<gco:CharacterString>ISO 19115-2 Geographic Information - Metadata Part 2 Extensions for imagery and gridded data</gco:CharacterString>
</gmd:metadataStandardName>
<gmd:metadataStandardVersion>
<gco:CharacterString>ISO 19115-2:2009(E)</gco:CharacterString>
</gmd:metadataStandardVersion>
<!-- This section documents granule data in this metadata record. -->
<gmd:identificationInfo>
<gmd:MD_DataIdentification>
<!-- This section holds the granule MetadataProviderDates, and the granule identifiers -->
<gmd:citation>
<gmd:CI_Citation>
<!-- The granule doesn't have a title -->
<gmd:title gco:nilReason="inapplicable"/>
<!-- The date/time that data provider created the granule info on data provider's database.-->
-->
<gmd:date>
<gmd:CI_Date>
<gmd:date>
<gco:DateTime>2021-07-29T19:16:54.670287Z</gco:DateTime>
</gmd:date>
<gmd:dateType>
<gmd:CI_DateTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#CI_DateTypeCode" codeListValue="creation">MISSING:DataDateTypeCode</gmd:CI_DateTypeCode>
</gmd:dateType>
</gmd:CI_Date>
</gmd:date>
<!-- This is the producer granule id -->
<gmd:identifier>
<gmd:MD_Identifier>
<gmd:code>
<gco:CharacterString>v1.0.0</gco:CharacterString>
</gmd:code>
<gmd:codeSpace>
<gco:CharacterString>gov.nasa.esdis.umm.otherid</gco:CharacterString>
</gmd:codeSpace>
<gmd:description>
<gco:CharacterString>OtherId: PGEVersionId</gco:CharacterString>
</gmd:description>
</gmd:MD_Identifier>
</gmd:identifier>
<gmd:otherCitationDetails>
<gco:CharacterString/>
</gmd:otherCitationDetails>
</gmd:CI_Citation>
</gmd:citation>
<gmd:abstract gco:nilReason="inapplicable"/>
<gmd:purpose gco:nilReason="inapplicable"/>
<!-- This secton holds the ReprocessingPlanned value -->
<gmd:resourceMaintenance>
<gmd:MD_MaintenanceInformation>
<gmd:maintenanceAndUpdateFrequency>
<gmd:MD_MaintenanceFrequencyCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#MD_MaintenanceFrequencyCode" codeListValue="MD_MaintenanceFrequencyCode_annually">"Annually"</gmd:MD_MaintenanceFrequencyCode>
</gmd:maintenanceAndUpdateFrequency>
<gmd:maintenanceNote>
<gco:CharacterString>"ReprocessingPlanned:" MISSING:DataReprocessingPlanNote</gco:CharacterString>
</gmd:maintenanceNote>
</gmd:MD_MaintenanceInformation>
</gmd:resourceMaintenance>
<!-- This is where Spatial/GranuleLocalities are described. -->
<gmd:descriptiveKeywords>
<gmd:MD_Keywords>
<gmd:keyword>
<gco:CharacterString>MISSING:GranuleLocality</gco:CharacterString>
</gmd:keyword>
<gmd:type>
<gmd:MD_KeywordTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#MD_KeywordTypeCode" codeListValue="place">place</gmd:MD_KeywordTypeCode>
</gmd:type>
</gmd:MD_Keywords>
</gmd:descriptiveKeywords>
<!-- This section describes projects as keywords. The CMR does not read this section.-->
<gmd:descriptiveKeywords>
</gmd:descriptiveKeywords>
<!-- This section describes platform keywords -->
<gmd:descriptiveKeywords>
</gmd:descriptiveKeywords>
<!-- This section describes instrument keywords -->
<gmd:descriptiveKeywords>
</gmd:descriptiveKeywords>
<!-- This section holds the granule access constraints. The AccessConstraintsValue can be used in the MMT (Metadata Management Tool) manage CMR section to manage granule level access control. -->
<!-- This is the granules collection short name. If this is used then the Collection Version must also exist. Only this and Collection Versionor or Collection Entry Id are required. -->
<gmd:aggregationInfo>
<gmd:MD_AggregateInformation>
<gmd:aggregateDataSetIdentifier>
<gmd:MD_Identifier>
<gmd:code>
<gco:CharacterString>MAIA_L1GRM</gco:CharacterString>
</gmd:code>
<gmd:codeSpace>
<gco:CharacterString>gov.nasa.esdis.umm.collectionshortname</gco:CharacterString>
</gmd:codeSpace>
<gmd:description>
<gco:CharacterString>CollectionShortName</gco:CharacterString>
</gmd:description>
</gmd:MD_Identifier>
</gmd:aggregateDataSetIdentifier>
<gmd:associationType>
<gmd:DS_AssociationTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#DS_AssociationTypeCode" codeListValue="LargerWorkCitation">LargerWorkCitation</gmd:DS_AssociationTypeCode>
</gmd:associationType>
</gmd:MD_AggregateInformation>
</gmd:aggregationInfo>
<!-- This is the granules collection version. If this is used then the Collection Short Name must also exist. Only this and Collection Short Name or Collection Entry Id are required. -->
<gmd:aggregationInfo>
<gmd:MD_AggregateInformation>
<gmd:aggregateDataSetIdentifier>
<gmd:MD_Identifier>
<gmd:code>
<gco:CharacterString>MISSING:CollectionVersion</gco:CharacterString>
</gmd:code>
<gmd:codeSpace>
<gco:CharacterString>gov.nasa.esdis.umm.collectionversion</gco:CharacterString>
</gmd:codeSpace>
<gmd:description>
<gco:CharacterString>CollectionVersion</gco:CharacterString>
</gmd:description>
</gmd:MD_Identifier>
</gmd:aggregateDataSetIdentifier>
<gmd:associationType>
<gmd:DS_AssociationTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#DS_AssociationTypeCode" codeListValue="LargerWorkCitation">LargerWorkCitation</gmd:DS_AssociationTypeCode>
</gmd:associationType>
</gmd:MD_AggregateInformation>
</gmd:aggregationInfo>
<!-- This is where View Related Information or Project Home Page RelatedUrls go.-->
<gmd:aggregationInfo>
<gmd:MD_AggregateInformation>
<gmd:aggregateDataSetName>
<gmd:CI_Citation>
<gmd:title/>
<gmd:date/>
<gmd:citedResponsibleParty>
<gmd:CI_ResponsibleParty>
<gmd:contactInfo>
<gmd:CI_Contact>
<gmd:onlineResource>
<gmd:CI_OnlineResource>
<gmd:linkage>
<gmd:URL>https://maia.jpl.nasa.gov/</gmd:URL>
</gmd:linkage>
<gmd:description>
<gco:CharacterString>The MAIA project website offers background on the MAIA project, instrument, and data products, as well as resources and FAQs.</gco:CharacterString>
</gmd:description>
<gmd:function>
<gmd:CI_OnLineFunctionCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#CI_OnLineFunctionCode" codeListValue="information">information</gmd:CI_OnLineFunctionCode>
</gmd:function>
</gmd:CI_OnlineResource>
</gmd:onlineResource>
</gmd:CI_Contact>
</gmd:contactInfo>
<gmd:role/>
</gmd:CI_ResponsibleParty>
</gmd:citedResponsibleParty>
</gmd:CI_Citation>
</gmd:aggregateDataSetName>
<gmd:associationType/>
</gmd:MD_AggregateInformation>
</gmd:aggregationInfo>
<!-- this is the language used in the granule -->
<gmd:language>
<gco:CharacterString>eng</gco:CharacterString>
</gmd:language>
<!-- this is the character set used in the granule -->
<gmd:characterSet>
<gmd:MD_CharacterSetCode codeList="http://www.ngdc.noaa.gov/metadata/published/xsd/schema/resources/Codelist/gmxCodelists.xml#MD_CharacterSetCode" codeListValue="utf8">utf8</gmd:MD_CharacterSetCode>
</gmd:characterSet>
<!-- This section documents the granules spatial and temporal extent as well as the grid mapping names and the projection names - NOT the TilingIdentificationSystem. -->
<gmd:extent>
<!-- the EX_Extent id must exist with boundingExtent -->
<gmd:EX_Extent id="boundingExtent">
<!--This section describes a spatial extent gpolygon. -->
<gmd:geographicElement>
<gmd:EX_BoundingPolygon>
<gmd:polygon>
<!-- the unique ID within the record must exist -->
<gml:Polygon gml:id="d11e78">
<gml:exterior>
<!-- latitude first then longitude for every point - no commas just spaces - so latitude1 longitude1 latitude2 longitude2 ...
The points must be in counter clockwise order and closed (first point must match the last point). -->
<gml:LinearRing>
<gml:posList>None</gml:posList>
</gml:LinearRing>
</gml:exterior>
</gml:Polygon>
</gmd:polygon>
</gmd:EX_BoundingPolygon>
</gmd:geographicElement>
<!--This section describes a spatial extent bounding rectangle. -->
<gmd:geographicElement>
<!--Bounding Rectangle-->
<gmd:EX_GeographicBoundingBox>
<gmd:westBoundLongitude>
<gco:Decimal>None</gco:Decimal>
</gmd:westBoundLongitude>
<gmd:eastBoundLongitude>
<gco:Decimal>None</gco:Decimal>
</gmd:eastBoundLongitude>
<gmd:southBoundLatitude>
<gco:Decimal>None</gco:Decimal>
</gmd:southBoundLatitude>
<gmd:northBoundLatitude>
<gco:Decimal>None</gco:Decimal>
</gmd:northBoundLatitude>
</gmd:EX_GeographicBoundingBox>
</gmd:geographicElement>
<!-- This section documents the NativeProjectionNames -->
<gmd:geographicElement>
<!-- Must use NativeProjectionName* where * = 1, 2, 3, 4, etc. -->
<!-- List of projections allowed for this section; ESDIS, Tuesday, November 6, 2018 at 1:26 PM
For the gridded product, L2B_LR_* products - it could be UTM
ESDIS: what do we populate here 1) Native sampling coordinate (radar instrument observation coordinate) for L1 products, 2) Gridded products L2B_LR_SSH? -->
<gmd:EX_GeographicDescription id="AlbersEqualAreaConic">
<gmd:geographicIdentifier>
<gmd:MD_Identifier>
<gmd:code>
<gco:CharacterString>AlbersEqualAreaConic</gco:CharacterString>
</gmd:code>
<gmd:codeSpace>
<gco:CharacterString>gov.nasa.esdis.umm.nativeprojectionname</gco:CharacterString>
</gmd:codeSpace>
<gmd:description>
<gco:CharacterString>NativeProjectionName</gco:CharacterString>
</gmd:description>
</gmd:MD_Identifier>
</gmd:geographicIdentifier>
</gmd:EX_GeographicDescription>
</gmd:geographicElement>
<!-- This is the granules temporal extent -->
<gmd:temporalElement>
<!--RangeDateTime-->
<gmd:EX_TemporalExtent id="standard-time-range">
<gmd:extent>
<gml:TimePeriod gml:id="3bb85f72-14ef">
<gml:beginPosition>2019-01-02</gml:beginPosition>
<gml:endPosition>2019-01-02</gml:endPosition>
</gml:TimePeriod>
</gmd:extent>
</gmd:EX_TemporalExtent>
</gmd:temporalElement>
<gmd:temporalElement>
<!--RangeDateTime-->
<gmd:EX_TemporalExtent id="local-time-zone-range">
<gmd:extent>
<gml:TimePeriod gml:id="74790353-0302">
<gml:beginPosition>MISSING:LocalTimeRangeBeginLocalDateTime</gml:beginPosition>
<gml:endPosition>MISSING:LocalTimeRangeEndLocalDateTime</gml:endPosition>
</gml:TimePeriod>
</gmd:extent>
</gmd:EX_TemporalExtent>
</gmd:temporalElement>
</gmd:EX_Extent>
</gmd:extent>
<gmd:supplementalInformation/>
</gmd:MD_DataIdentification>
</gmd:identificationInfo>
<!-- This is the Measured Parameters section - it needs to be in its own contentInfo section - not within Additional Attributes, DayNightFlag, or CloudCover -->
<gmd:contentInfo>
<gmd:MD_CoverageDescription>
<gmd:attributeDescription>
<gco:RecordType>MeasuredParameters</gco:RecordType>
</gmd:attributeDescription>
<gmd:contentType>
<gmd:MD_CoverageContentTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#MD_CoverageContentTypeCode" codeListValue="physicalMeasurement">physicalMeasurement</gmd:MD_CoverageContentTypeCode>
</gmd:contentType>
<gmd:dimension>
<gmd:MD_Band>
<gmd:sequenceIdentifier>
<gco:MemberName>
<gco:aName>
<gco:CharacterString>MISSING:ParameterName</gco:CharacterString>
</gco:aName>
<gco:attributeType>
<gco:TypeName>
<gco:aName>
<gco:CharacterString>MeasuredParameters</gco:CharacterString>
</gco:aName>
</gco:TypeName>
</gco:attributeType>
</gco:MemberName>
</gmd:sequenceIdentifier>
<gmd:otherProperty>
<gco:Record>
<eos:AdditionalAttributes>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="qualityInformation">qualityInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>QAPercentMissingData</gco:CharacterString>
</eos:name>
<eos:dataType>
<eos:EOS_AdditionalAttributeDataTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeDataTypeCode" codeListValue="float">float</eos:EOS_AdditionalAttributeDataTypeCode>
</eos:dataType>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:QAPercentMissingData</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
</eos:AdditionalAttributes>
</gco:Record>
</gmd:otherProperty>
</gmd:MD_Band>
</gmd:dimension>
</gmd:MD_CoverageDescription>
</gmd:contentInfo>
<!-- This is the data quality section. It holds ReprocessingActual, ProductionDateTime, PGEVersionClass, Some AdditionalAttributes, and InputGranules. The MeasuredParameters go into its own dataQualityInfo Section. -->
<gmd:dataQualityInfo>
<gmd:DQ_DataQuality>
<!-- this lists that the scope for the data quality section pertains to the data set - the granule. -->
<gmd:scope>
<gmd:DQ_Scope>
<gmd:level>
<gmd:MD_ScopeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/gmxCodelists.xml#MD_ScopeCode" codeListValue="dataset">dataset</gmd:MD_ScopeCode>
</gmd:level>
</gmd:DQ_Scope>
</gmd:scope>
<gmd:lineage>
<gmd:LI_Lineage>
<gmd:processStep>
<gmi:LE_ProcessStep>
<gmd:description>
<gco:CharacterString>ReprocessingActual: MISSING:MAIA_ReprocessingActualStatement</gco:CharacterString>
</gmd:description>
</gmi:LE_ProcessStep>
</gmd:processStep>
<!-- This is the production date time -->
<gmd:processStep>
<gmi:LE_ProcessStep>
<gmd:description>
<gco:CharacterString>ProductionDateTime</gco:CharacterString>
</gmd:description>
<gmd:dateTime>
<gco:DateTime>2021-07-29T19:16:54.670287Z</gco:DateTime>
</gmd:dateTime>
</gmi:LE_ProcessStep>
</gmd:processStep>
<!-- This section holds the GranuleInputs -->
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule1">MISSING:InputGranule1</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule2">MISSING:InputGranule2</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule3">MISSING:InputGranule3</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule4">MISSING:InputGranule4</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule5">MISSING:InputGranule5</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule6">MISSING:InputGranule6</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule7">MISSING:InputGranule7</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule8">MISSING:InputGranule8</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule9">MISSING:InputGranule9</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule10">MISSING:InputGranule10</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule11">MISSING:InputGranule11</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule12">MISSING:InputGranule12</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule13">MISSING:InputGranule13</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule14">MISSING:InputGranule14</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule15">MISSING:InputGranule15</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule16">MISSING:InputGranule16</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule17">MISSING:InputGranule17</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule18">MISSING:InputGranule18</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule19">MISSING:InputGranule19</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule20">MISSING:InputGranule20</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule21">MISSING:InputGranule21</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule22">MISSING:InputGranule22</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule23">MISSING:InputGranule23</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule24">MISSING:InputGranule24</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule25">MISSING:InputGranule25</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule26">MISSING:InputGranule26</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule27">MISSING:InputGranule27</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule28">MISSING:InputGranule28</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule29">MISSING:InputGranule29</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule30">MISSING:InputGranule30</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule31">MISSING:InputGranule31</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule32">MISSING:InputGranule32</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule33">MISSING:InputGranule33</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule34">MISSING:InputGranule34</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule35">MISSING:InputGranule35</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule36">MISSING:InputGranule36</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule37">MISSING:InputGranule37</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule38">MISSING:InputGranule38</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule39">MISSING:InputGranule39</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule40">MISSING:InputGranule40</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule41">MISSING:InputGranule41</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule42">MISSING:InputGranule42</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule43">MISSING:InputGranule43</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule44">MISSING:InputGranule44</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule45">MISSING:InputGranule45</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule46">MISSING:InputGranule46</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule47">MISSING:InputGranule47</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule48">MISSING:InputGranule48</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule49">MISSING:InputGranule49</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
<gmd:source>
<gmi:LE_Source>
<gmd:description>
<gco:CharacterString>GranuleInput</gco:CharacterString>
</gmd:description>
<gmd:sourceCitation>
<gmd:CI_Citation>
<gmd:title>
<gmx:FileName src="MISSING:InputGranule50">MISSING:InputGranule50</gmx:FileName>
</gmd:title>
<gmd:date gco:nilReason="unknown"/>
</gmd:CI_Citation>
</gmd:sourceCitation>
</gmi:LE_Source>
</gmd:source>
</gmd:LI_Lineage>
</gmd:lineage>
</gmd:DQ_DataQuality>
</gmd:dataQualityInfo>
<!-- This section describes the Platform, Instrument, and Project information -->
<gmi:acquisitionInformation>
<gmi:MI_AcquisitionInformation>
<!-- This section describes the Platform and an embedded Instrument -->
<gmi:platform>
<eos:EOS_Platform id="MAIA">
<gmi:identifier>
<gmd:MD_Identifier>
<gmd:code>
<gco:CharacterString>MAIA</gco:CharacterString>
</gmd:code>
<gmd:codeSpace>
<gco:CharacterString>gov.nasa.esdis.umm.platformshortname</gco:CharacterString>
</gmd:codeSpace>
<gmd:description>
<gco:CharacterString>PlatformShortName</gco:CharacterString>
</gmd:description>
</gmd:MD_Identifier>
</gmi:identifier>
<gmi:description gco:nilReason="missing"/>
<!-- the xlink:href= is not needed if the instrument in embedded with in the platform. It is necessary if the instrument is not embedded -->
<!-- Use this section for all instrument characteristics-->
<gmi:instrument xlink:href="MAIA">
<eos:EOS_Instrument id="MAIA">
<gmi:identifier>
<gmd:MD_Identifier>
<gmd:code>
<gco:CharacterString>MAIA</gco:CharacterString>
</gmd:code>
<gmd:codeSpace>
<gco:CharacterString>gov.nasa.esdis.umm.instrumentshortname</gco:CharacterString>
</gmd:codeSpace>
<gmd:description>
<gco:CharacterString>InstrumentShortName</gco:CharacterString>
</gmd:description>
</gmd:MD_Identifier>
</gmi:identifier>
<gmi:type gco:nilReason="inapplicable"/>
<gmi:mountedOn xlink:href="MAIA"/>
<!--Instrument Characteristics and operational modes-->
<eos:otherProperty>
<gco:Record>
<eos:AdditionalAttribute>
<!-- This next 2 sections are the instruments characteristics -->
<!-- TODO: Capture any high level instrument characteristics such as band & bandwidth, etc -->
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="instrumentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString></gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString></gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="instrumentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>NumberOfInputViews</gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:NumberOfInputViews</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="contentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>TargetType</gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:TargetType</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="contentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>Total_PM2.5_MedianValue</gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:TotalPM25_MedianValue</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="contentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>Total_PM2.5_Max_Value</gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:TotalPM25_MaxValue</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="contentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>Total_PM10_MedianValue</gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:TotalPM10_MedianValue</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="contentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>Total_PM10_Max_Value</gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:TotalPM10_MaxValue</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
<eos:AdditionalAttribute>
<eos:reference>
<eos:EOS_AdditionalAttributeDescription>
<eos:type>
<eos:EOS_AdditionalAttributeTypeCode codeList="https://cdn.earthdata.nasa.gov/iso/resources/Codelist/eosCodelists.xml#EOS_AdditionalAttributeTypeCode" codeListValue="contentInformation">instrumentInformation</eos:EOS_AdditionalAttributeTypeCode>
</eos:type>
<eos:name>
<gco:CharacterString>DayOfWeek</gco:CharacterString>
</eos:name>
</eos:EOS_AdditionalAttributeDescription>
</eos:reference>
<eos:value>
<gco:CharacterString>MISSING:DayOfWeek</gco:CharacterString>
</eos:value>
</eos:AdditionalAttribute>
</eos:AdditionalAttribute>
</gco:Record>
</eos:otherProperty>
</eos:EOS_Instrument>
</gmi:instrument>
</eos:EOS_Platform>
</gmi:platform>
</gmi:MI_AcquisitionInformation>
</gmi:acquisitionInformation>
</gmi:MI_Metadata>
product_version = v1.0.0
software_version = v1.0.0
data_version = v1.0.0
start_date = 2019-01-02
end_date = 2019-01-02
production_time = 2021-07-29T19:16:54.670287Z
production_hostname = 36ff6f07bfe6Read the netCDF Variables and Attribute Metadata
var_list = []
var_attr_list = []
var_data_list = []
def readVars ( nc_file ):
vars = nc_file.variables.keys()
for var_name in vars:
attr = nc_file.variables[var_name]
vardata = nc_file.variables[var_name][:]
var_attr_list.append( attr )
var_data_list.append( vardata)
return vars, var_attr_list, var_data_listRead and Describe the netCDF Un-Grouped Variable Attributes
print ('Un-Grouped netCDF Variables:')
[vars, var_attr_list, var_data_list] = readVars( nc_file )
nvars = len(vars)
print ('Number of Un-Grouped variables = ', nvars)
for i in range(0, nvars):
vardata = var_data_list[i]
print ('----------')
print (var_attr_list[i])
print (list(vars)[i], '[0:10] =\n', list(vardata)[0:10])Un-Grouped netCDF Variables:
Number of Un-Grouped variables = 3
----------
<class 'netCDF4._netCDF4.Variable'>
float32 X_Dim(X_Dim)
_FillValue: 9.96921e+36
unit: meter
standard_name: projection_x_coordinate
long_name: Projection X Coordinate
_CoordinateAxisType: GeoX
unlimited dimensions: X_Dim
current shape = (352,)
filling on
X_Dim [0:10] =
[-175625.0, -174625.0, -173625.0, -172625.0, -171625.0, -170625.0, -169625.0, -168625.0, -167625.0, -166625.0]
----------
<class 'netCDF4._netCDF4.Variable'>
float32 Y_Dim(Y_Dim)
_FillValue: 9.96921e+36
unit: meter
standard_name: projection_y_coordinate
long_name: Projection Y Coordinate
_CoordinateAxisType: GeoY
unlimited dimensions: Y_Dim
current shape = (420,)
filling on
Y_Dim [0:10] =
[209625.0, 208625.0, 207625.0, 206625.0, 205625.0, 204625.0, 203625.0, 202625.0, 201625.0, 200625.0]
----------
<class 'netCDF4._netCDF4.Variable'>
int32 Albers_Equal_Area()
semi_major_axis: 6378137.0
inverse_flattening: 298.257223563
min_x: -500000.0
min_y: -500000.0
max_x: 500000.0
max_y: 500000.0
long_name: Albers Equal Area Conic Projection
grid_mapping_name: albers_conical_equal_area
standard_parallel: [33.3 37.3]
longitude_of_central_meridian: -118.6
latitude_of_projection_origin: 35.3
false_easting: 0.0
false_northing: 0.0
origin_code: 0
pix_reg_code: 0
crs_wkt: PROJCS["unnamed",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0],UNIT["degree",0.0174532925199433],AUTHORITY["EPSG","4326"]],PROJECTION["Albers_Conic_Equal_Area"],PARAMETER["standard_parallel_1",33.3],PARAMETER["standard_parallel_2",37.3],PARAMETER["latitude_of_center",35.3],PARAMETER["longitude_of_center",-118.6],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]]]
dimension_order: 1
unlimited dimensions:
current shape = ()
filling on, default _FillValue of -2147483647 used--------------------------------------------------------------------------- TypeError Traceback (most recent call last) <ipython-input-6-0d601eea16a1> in <module> 7 print ('----------') 8 print (var_attr_list[i]) ----> 9 print (list(vars)[i], '[0:10] =\n', list(vardata)[0:10]) TypeError: iteration over a 0-d array
List the Un-grouped Variables
list(vars)['X_Dim', 'Y_Dim', 'Albers_Equal_Area']Read the netCDF Group Variables
nc_file.groups{'Geometric_Parameters': <class 'netCDF4._netCDF4.Group'>
group /Geometric_Parameters:
dimensions(sizes):
variables(dimensions): float32 Longitude(X_Dim, Y_Dim), float32 Latitude(X_Dim, Y_Dim)
groups: ,
'PM_10': <class 'netCDF4._netCDF4.Group'>
group /PM_10:
dimensions(sizes):
variables(dimensions): float32 PM_10_Total_Mean(X_Dim, Y_Dim), float32 PM_10_Total_Uncertainty(X_Dim, Y_Dim)
groups: ,
'PM_2.5': <class 'netCDF4._netCDF4.Group'>
group /PM_2.5:
dimensions(sizes):
variables(dimensions): float32 PM_2.5_Dust_Mean(X_Dim, Y_Dim), float32 PM_2.5_Dust_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_EC_Mean(X_Dim, Y_Dim), float32 PM_2.5_EC_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_Nitrate_Mean(X_Dim, Y_Dim), float32 PM_2.5_Nitrate_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_OC_Mean(X_Dim, Y_Dim), float32 PM_2.5_OC_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_Sulfate_Mean(X_Dim, Y_Dim), float32 PM_2.5_Sulfate_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_Total_Mean(X_Dim, Y_Dim), float32 PM_2.5_Total_Uncertainty(X_Dim, Y_Dim)
groups: }Read the Geometric Parameters Group Variables
grp_geo = nc_file.groups['Geometric_Parameters']Describe the Geometric Group Variables
grp_geo<class 'netCDF4._netCDF4.Group'>
group /Geometric_Parameters:
dimensions(sizes):
variables(dimensions): float32 Longitude(X_Dim, Y_Dim), float32 Latitude(X_Dim, Y_Dim)
groups: List the Geometric Group Variables
list(grp_geo.variables)['Longitude', 'Latitude']Read the PM 10 Group Variables
grp_pm10 = nc_file.groups['PM_10']Describe the PM 10 Variables
grp_pm10<class 'netCDF4._netCDF4.Group'>
group /PM_10:
dimensions(sizes):
variables(dimensions): float32 PM_10_Total_Mean(X_Dim, Y_Dim), float32 PM_10_Total_Uncertainty(X_Dim, Y_Dim)
groups: List the PM 10 Variables
list(grp_pm10.variables)['PM_10_Total_Mean', 'PM_10_Total_Uncertainty']Read the PM 2.5 Group Variables
grp_pm2d5 = nc_file.groups['PM_2.5']Describe the PM 2.5 Variables
grp_pm2d5<class 'netCDF4._netCDF4.Group'>
group /PM_2.5:
dimensions(sizes):
variables(dimensions): float32 PM_2.5_Dust_Mean(X_Dim, Y_Dim), float32 PM_2.5_Dust_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_EC_Mean(X_Dim, Y_Dim), float32 PM_2.5_EC_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_Nitrate_Mean(X_Dim, Y_Dim), float32 PM_2.5_Nitrate_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_OC_Mean(X_Dim, Y_Dim), float32 PM_2.5_OC_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_Sulfate_Mean(X_Dim, Y_Dim), float32 PM_2.5_Sulfate_Uncertainty(X_Dim, Y_Dim), float32 PM_2.5_Total_Mean(X_Dim, Y_Dim), float32 PM_2.5_Total_Uncertainty(X_Dim, Y_Dim)
groups: List the PM 2.5 Variables
list(grp_pm2d5.variables)['PM_2.5_Dust_Mean',
'PM_2.5_Dust_Uncertainty',
'PM_2.5_EC_Mean',
'PM_2.5_EC_Uncertainty',
'PM_2.5_Nitrate_Mean',
'PM_2.5_Nitrate_Uncertainty',
'PM_2.5_OC_Mean',
'PM_2.5_OC_Uncertainty',
'PM_2.5_Sulfate_Mean',
'PM_2.5_Sulfate_Uncertainty',
'PM_2.5_Total_Mean',
'PM_2.5_Total_Uncertainty']Reading the Discrete Variables within the Variable Groups
lon = grp_geo.variables['Longitude'][:]
lat = grp_geo.variables['Latitude'][:]
pm_10_total = grp_pm10.variables['PM_10_Total_Mean'][:]
pm_10_uncert = grp_pm10.variables['PM_10_Total_Uncertainty'][:]print, lon(<function print>,
masked_array(
data=[[-120.57638 , -120.57616 , -120.575935, ..., -120.487076,
-120.48687 , -120.486664],
[-120.565125, -120.5649 , -120.564674, ..., -120.47632 ,
-120.47612 , -120.475914],
[-120.55386 , -120.55365 , -120.55342 , ..., -120.465576,
-120.46537 , -120.465164],
...,
[-116.64613 , -116.646355, -116.64658 , ..., -116.73443 ,
-116.734634, -116.73483 ],
[-116.63488 , -116.6351 , -116.63533 , ..., -116.72368 ,
-116.723885, -116.72409 ],
[-116.62363 , -116.62384 , -116.62407 , ..., -116.71293 ,
-116.713135, -116.71334 ]],
mask=False,
fill_value=1e+20,
dtype=float32))print, lat(<function print>,
masked_array(
data=[[37.171444, 37.162437, 37.153427, ..., 33.415184, 33.40617 ,
33.397156],
[37.171623, 37.162617, 37.153606, ..., 33.415356, 33.40634 ,
33.397327],
[37.171803, 37.162792, 37.153786, ..., 33.415524, 33.40651 ,
33.397495],
...,
[37.171803, 37.162792, 37.153786, ..., 33.415524, 33.40651 ,
33.397495],
[37.171623, 37.162617, 37.153606, ..., 33.415356, 33.40634 ,
33.397327],
[37.171444, 37.162437, 37.153427, ..., 33.415184, 33.40617 ,
33.397156]],
mask=False,
fill_value=1e+20,
dtype=float32))Generate a “Psuedo” AlbersEqualArea 2-D Lat/Lon Map of PM 10 Total Mean
plt.figure(figsize=(10, 10))
ax1 = plt.axes(projection=ccrs.AlbersEqualArea())
ax1.gridlines(crs=ccrs.AlbersEqualArea(central_longitude=-118.6, central_latitude=35.3, false_easting=0.0, false_northing=0.0, standard_parallels=(33.3, 37.3), globe=None), alpha=0.3)
ax1.coastlines()
plt.figure(figsize=(10, 10))
ax2 = plt.axes(projection=ccrs.PlateCarree())
ax2.gridlines(crs=ccrs.PlateCarree(), draw_labels=True, alpha=0.3)
ax2.coastlines()
x = ax2.scatter(lon[:], lat[:], c=pm_10_total[:], lw=2, vmin=35., vmax=40.)
plt.colorbar(x, orientation='horizontal', pad=0.05)
plt.show()
print('Units = micrograms m^-3')

Units = micrograms m^-3Generate PlateCarree 2-D Lat/Lon Map of PM 10 Total Mean
plt.figure(figsize=(10, 10))
ax1 = plt.axes(projection=ccrs.PlateCarree())
ax1.gridlines(crs=ccrs.PlateCarree(), draw_labels=True, alpha=0.3)
ax1.coastlines()
plt.figure(figsize=(10, 10))
ax2 = plt.axes(projection=ccrs.PlateCarree())
ax2.gridlines(crs=ccrs.PlateCarree(), draw_labels=True, alpha=0.3)
ax2.coastlines()
x = ax2.scatter(lon[:], lat[:], c=pm_10_total[:], lw=2, vmin=35., vmax=40.)
plt.colorbar(x, orientation='horizontal', pad=0.05)
plt.show()
print('Units = micrograms m^-3')

Units = micrograms m^-3Generate a 2-D Lat/Lon Map of PM 10 Total Uncertainty
plt.figure(figsize=(10, 10))
ax1 = plt.axes(projection=ccrs.PlateCarree())
ax1.gridlines(crs=ccrs.PlateCarree(), draw_labels=True, alpha=0.3)
ax1.coastlines()
plt.figure(figsize=(10, 10))
ax2 = plt.axes(projection=ccrs.PlateCarree())
ax2.gridlines(crs=ccrs.PlateCarree(), draw_labels=True, alpha=0.3)
ax2.coastlines()
x = ax2.scatter(lon[:], lat[:], c=pm_10_uncert[:], lw=2, vmin=7.5, vmax=9.)
plt.colorbar(x, orientation='horizontal', pad=0.05)
plt.show()
print('Units = micrograms m^-3')

Units = micrograms m^-3