Ingest Browse Generation
This entry documents how to setup a workflow that utilizes Cumulus's built-in granule file type configuration such that on ingest the browse data is exported to CMR.
We will discuss how to run a processing workflow against an inbound granule that has data but no browse generated. The workflow will generate a browse file and add the appropriate output values to the Cumulus message so that the built-in post-to-cmr task will publish the data appropriately.
Sections
Prerequisites
Cumulus
This entry assumes you have a deployed instance of Cumulus v1.16.0 or later, and a working dashboard following the instructions in the deployment documentation. This entry also assumes you have some knowledge of how to configure Collections, Providers and Rules and basic Cumulus operation.
Prior to working through this entry, you should be somewhat familiar with the Hello World example the Workflows section of the documentation, and building Cumulus lambdas.
You should also review the Data Cookbooks Setup portion of the documentation as it contains useful information on the inter-task message schema expectations.
This entry will utilize the dashboard application. You will need to have a dashboard deployed as described in the Cumulus deployment documentation to follow the instructions in this example.
If you'd prefer to not utilize a running dashboard to add Collections, Providers and trigger Rules, you can set the Collection/Provider and Rule via the API, however in that instance you should be very familiar with the Cumulus API before attempting the example in this entry.
Common Metadata Repository
You should be familiar with the Common Metadata Repository and already be set up as a provider with configured collections and credentials to ingest data into CMR. You should know what the collection name and version number are.
Source Data
You should have data available for Cumulus to ingest in an S3 bucket that matches with CMR if you'd like to push a record to CMR UAT.
For the purposes of this entry, we will be using a pre-configured MOD09GQ version 006 CMR collection. If you'd prefer to utilize the example processing code, using mocked up data files matching the file naming convention will suffice, so long as you also have a matching collection setup in CMR.
If you'd prefer to ingest another data type, you will need to generate a processing lambda (see Build Processing Lambda below).
Configure Cumulus
CMR
To run this example with successful exports to CMR you'll need to make sure the CMR configuration keys for the cumulus terraform module are configured per that module's variables.tf file.
Workflows
Summary
For this example, you are going to be adding two workflows to your Cumulus deployment.
DiscoverGranulesBrowseExample
This workflow will run the
DiscoverGranulestask, targeting the S3 bucket/folder mentioned in the prerequisites. The output of that task will be passed into QueueGranules, which will trigger the second workflow for each granule to be ingested. The example presented here will be a single granule with a .hdf data file and a .met metadata file only, however your setup may result in more granules, or different files.CookbookBrowseExample
This workflow will be triggered for each granule in the previous workflow. It will utilize the SyncGranule task, which brings the files into a staging location in the Cumulus buckets.
The output from this task will be passed into the
ProcessingStepstep , which in this example will utilize theFakeProcessingLambdatask we provide for testing/as an example in Core, however to use your own data you will need to write a lambda that generates the appropriate CMR metadata file and accepts and returns appropriate task inputs and outputs.From that task we will utilize a core task
FilesToGranulesthat will transform the processing output event.input list/config.InputGranules into an array of Cumulus granules objects.Using the generated granules list, we will utilize the core task
MoveGranulesto move the granules to the target buckets as defined in the collection configuration. That task will transfer the files to their final storage location and update the CMR metadata files and the granules list as output.That output will be used in the
PostToCmrtask combined with the previously generated CMR file to export the granule metadata to CMR.
Workflow Configuration
Copy the following workflow deployment files to your deployment's main directory (including the referenced .asl.json files):
browse_example.tf(DiscoverGranulesBrowseExampleworkflow)cookbook_browse_example_workflow.tf(CookbookBrowseExampleworkflow)
Please Note: You should update the source = line to match the current Cumulus workflow module release artifact to the version of Cumulus you're deploying:
source = "https://github.com/nasa/cumulus/releases/download/{version}/terraform-aws-cumulus-workflow.zip"
A few things to note about tasks in the workflow being added:
Note: In the snippets below,
${post_to_cmr_task_arn}and${fake_processing_task_arn}are interpolated values referring to Terraform resources. See the example deployment code for theCookbookBrowseExampleworkflow.
- The CMR step in CookbookBrowseExample:
"CmrStep": {
"Parameters": {
"cma": {
"event.$": "$",
"task_config": {
"bucket": "{$.meta.buckets.internal.name}",
"stack": "{$.meta.stack}",
"cmr": "{$.meta.cmr}",
"launchpad": "{$.meta.launchpad}",
"input_granules": "{$.meta.input_granules}",
"granuleIdExtraction": "{$.meta.collection.granuleIdExtraction}"
}
}
},
"Type": "Task",
"Resource": "${post_to_cmr_task_arn}",
"Retry": [
{
"ErrorEquals": [
"Lambda.ServiceException",
"Lambda.AWSLambdaException",
"Lambda.SdkClientException"
],
"IntervalSeconds": 2,
"MaxAttempts": 6,
"BackoffRate": 2
}
],
"Catch": [
{
"ErrorEquals": [
"States.ALL"
],
"ResultPath": "$.exception",
"Next": "WorkflowFailed"
}
],
"End": true
}
Note that, in the task, the CmrStep.Parameters.cma.task_config.cmr key will contain the values you configured in the cmr configuration section above.
- The Processing step in CookbookBrowseExample:
"ProcessingStep": {
"Parameters": {
"cma": {
"event.$": "$",
"task_config": {
"bucket": "{$.meta.buckets.internal.name}",
"collection": "{$.meta.collection}",
"cmrMetadataFormat": "{$.meta.cmrMetadataFormat}",
"additionalUrls": "{$.meta.additionalUrls}",
"generateFakeBrowse": true,
"cumulus_message": {
"outputs": [
{
"source": "{$.granules}",
"destination": "{$.meta.input_granules}"
},
{
"source": "{$.files}",
"destination": "{$.payload}"
}
]
}
}
}
},
"Type": "Task",
"Resource": "${fake_processing_task_arn}",
"Catch": [
{
"ErrorEquals": [
"States.ALL"
],
"ResultPath": "$.exception",
"Next": "WorkflowFailed"
}
],
"Retry": [
{
"ErrorEquals": [
"States.ALL"
],
"IntervalSeconds": 2,
"MaxAttempts": 3
}
],
"Next": "FilesToGranulesStep"
},
If you're not ingesting mock data matching the example, or would like to use modify the example to ingest your own data please see the build-lambda section below. You will need to configure a different lambda entry for your lambda and utilize it in place of the Resource defined in the example workflow.
Please note: FakeProcessing is the core provided browse/CMR generation lambda we're using for the example in this entry.
Lambdas
All lambdas utilized in this example are provided in a standard deployment of Cumulus and require no additional configuration.
Redeploy
Once you've configured your CMR credentials, updated your workflow configuration, and updated your lambda configuration you should be able to redeploy your cumulus instance by running the following commands:
terraform init
You should expect to see output similar to:
$ terraform init
Initializing modules...
Downloading https://github.com/nasa/cumulus/releases/download/{version}/terraform-aws-cumulus-workflow.zip for cookbook_browse_example_workflow...
- cookbook_browse_example_workflow in .terraform/modules/cookbook_browse_example_workflow
Downloading https://github.com/nasa/cumulus/releases/download/{version}/terraform-aws-cumulus-workflow.zip for discover_granules_browse_example_workflow...
- discover_granules_browse_example_workflow in .terraform/modules/discover_granules_browse_example_workflow
Initializing the backend...
Initializing provider plugins...
Terraform has been successfully initialized!
You may now begin working with Terraform. Try running "terraform plan" to see
any changes that are required for your infrastructure. All Terraform commands
should now work.
If you ever set or change modules or backend configuration for Terraform,
rerun this command to reinitialize your working directory. If you forget, other
commands will detect it and remind you to do so if necessary.
terraform apply
You should expect to see output similar to the following truncated example:
$ terraform apply
module.cumulus.module.archive.null_resource.rsa_keys: Refreshing state... [id=xxxxxxxxx]
data.terraform_remote_state.data_persistence: Refreshing state...
module.cumulus.module.archive.aws_cloudwatch_event_rule.daily_execution_payload_cleanup: Refreshing state... [id=xxxx]
....
An execution plan has been generated and is shown below.
Resource actions are indicated with the following symbols:
+ create
~ update in-place
-/+ destroy and then create replacement
<= read (data resources)
Terraform will perform the following actions:
{...}
Plan: 15 to add, 3 to change, 1 to destroy.
Do you want to perform these actions?
Terraform will perform the actions described above.
Only 'yes' will be accepted to approve.
Enter a value: yes
{...}
Apply complete! Resources: 15 added, 3 changed, 1 destroyed.
Releasing state lock. This may take a few moments...
Outputs:
archive_api_redirect_uri = {URL}
archive_api_uri = {URL}
distribution_redirect_uri = {URL}
distribution_url = {URL}
s3_credentials_redirect_uri = {URL}
Configure Ingest
Now that Cumulus has been updated updated with the new workflows and code, we will use the Cumulus dashboard to configure an ingest collection, provider and rule so that we can trigger the configured workflow.
Add Collection
Navigate to the 'Collection' tab on the interface and add a collection.
{
"name": "MOD09GQ",
"version": "006",
"process": "modis",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{substring(file.name, 0, 3)}",
"duplicateHandling": "replace",
"granuleId": "^MOD09GQ\\.A[\\d]{7}\\.[\\S]{6}\\.006\\.[\\d]{13}$",
"granuleIdExtraction": "(MOD09GQ\\..*)(\\.hdf|\\.cmr|_ndvi\\.jpg|\\.jpg)",
"sampleFileName": "MOD09GQ.A2017025.h21v00.006.2017034065104.hdf",
"files": [
{
"bucket": "protected",
"regex": "^MOD09GQ\\.A[\\d]{7}\\.[\\S]{6}\\.006\\.[\\d]{13}\\.hdf$",
"sampleFileName": "MOD09GQ.A2017025.h21v00.006.2017034065104.hdf",
"type": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{extractYear(cmrMetadata.Granule.Temporal.RangeDateTime.BeginningDateTime)}/{substring(file.name, 0, 3)}"
},
{
"bucket": "private",
"regex": "^MOD09GQ\\.A[\\d]{7}\\.[\\S]{6}\\.006\\.[\\d]{13}\\.hdf\\.met$",
"sampleFileName": "MOD09GQ.A2017025.h21v00.006.2017034065104.hdf.met",
"type": "metadata"
},
{
"bucket": "protected-2",
"regex": "^MOD09GQ\\.A[\\d]{7}\\.[\\S]{6}\\.006\\.[\\d]{13}\\.cmr\\.xml$",
"sampleFileName": "MOD09GQ.A2017025.h21v00.006.2017034065104.cmr.xml"
},
{
"bucket": "protected",
"regex": "^MOD09GQ\\.A[\\d]{7}\\.[\\S]{6}\\.006\\.[\\d]{13}\\.jpg$",
"sampleFileName": "MOD09GQ.A2017025.h21v00.006.2017034065104.jpg"
}
]
}
Please note: Even though our initial discover granules ingest brings in only the .hdf and .met files we've staged, we still configure the other possible file types for this collection's granules.
Add Provider
Next navigate to the Provider tab and create a provider with the following values, using whatever name you wish, and the bucket the data was staged to as the host:
Name:
Protocol: S3
Host: {{data_source_bucket}}
Add Rule
Once you have your provider added, go to the Rules tab and add a rule with the following values (using whatever name you wish, populating the workflow and provider keys with the previously entered values) Note that you need to set the "provider_path" to the path on your bucket (e.g. "/data") that you've staged your mock/test data.:
{
"name": "TestBrowseGeneration",
"workflow": "DiscoverGranulesBrowseExample",
"provider": "{{provider_from_previous_step}}",
"collection": {
"name": "MOD09GQ",
"version": "006"
},
"meta": {
"provider_path": "{{path_to_data}}"
},
"rule": {
"type": "onetime"
},
"state": "ENABLED",
"updatedAt": 1553053438767
}
Run Workflows
Once you've configured the Collection and Provider and added a onetime rule, you're ready to trigger your rule, and watch the ingest workflows process.
Go to the Rules tab, click the rule you just created:
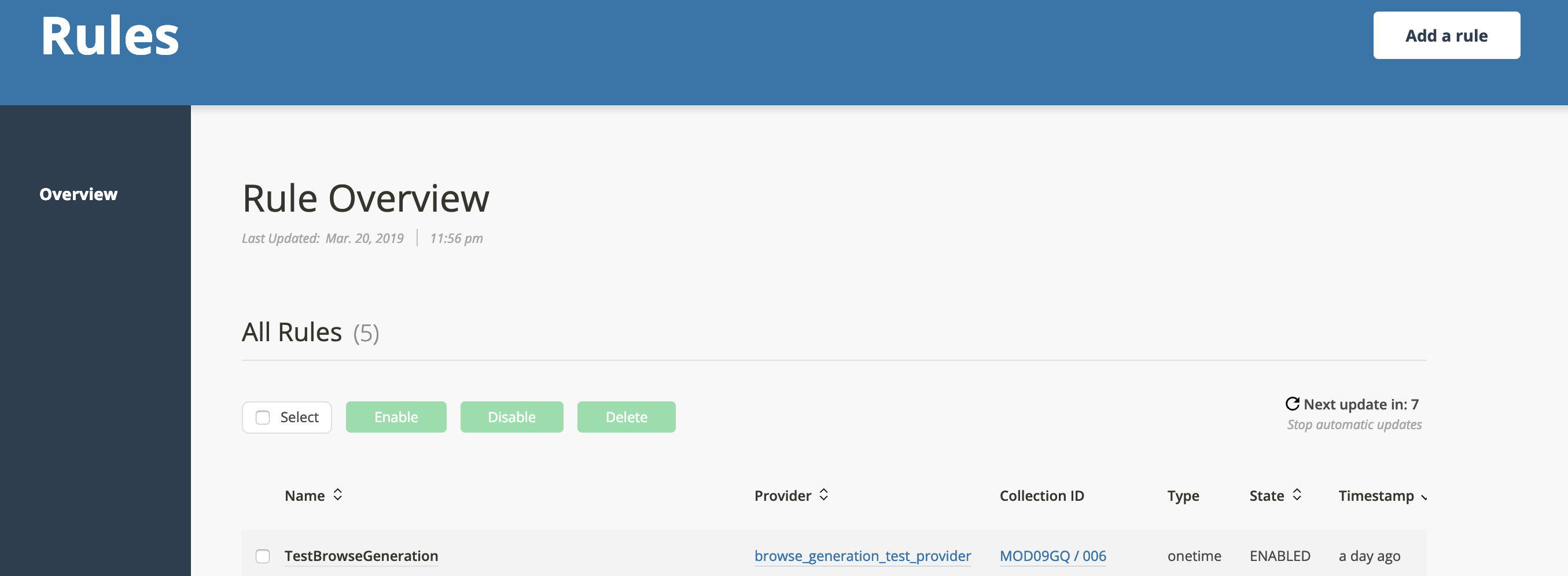
Then click the gear in the upper right corner and click "Rerun":
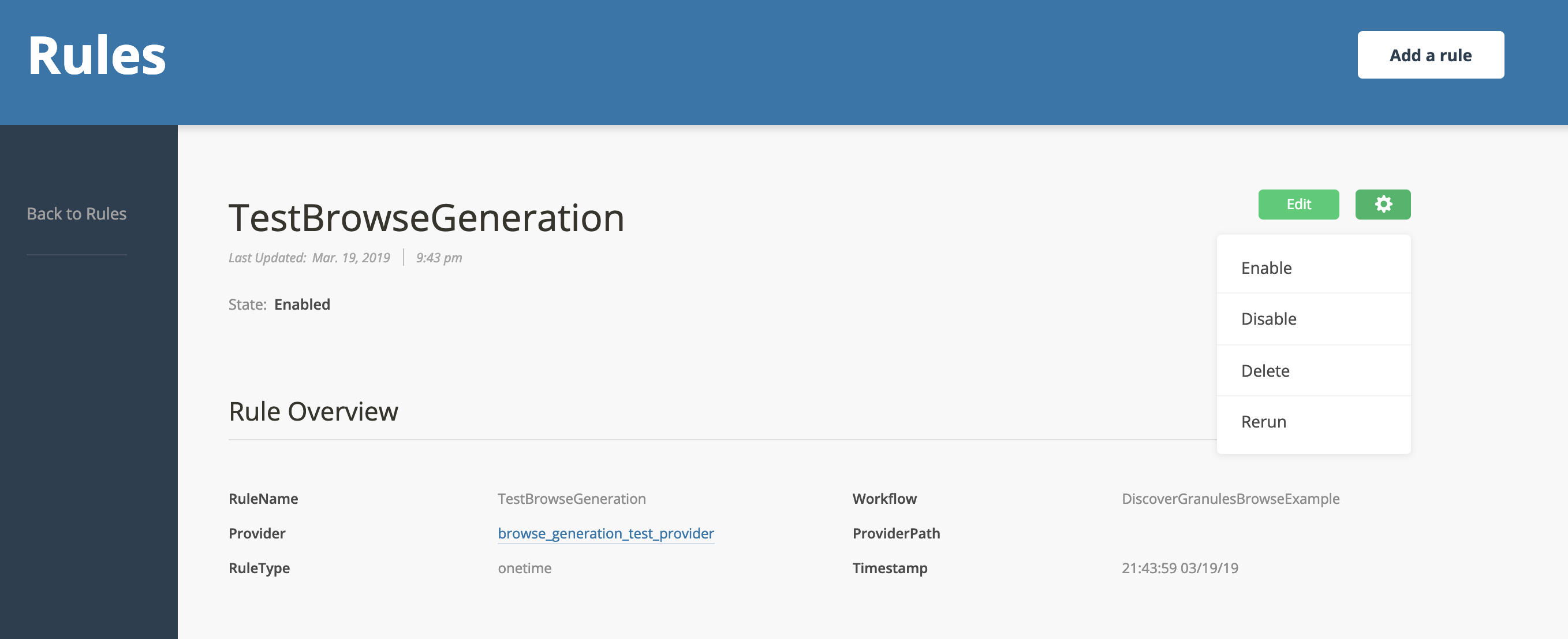
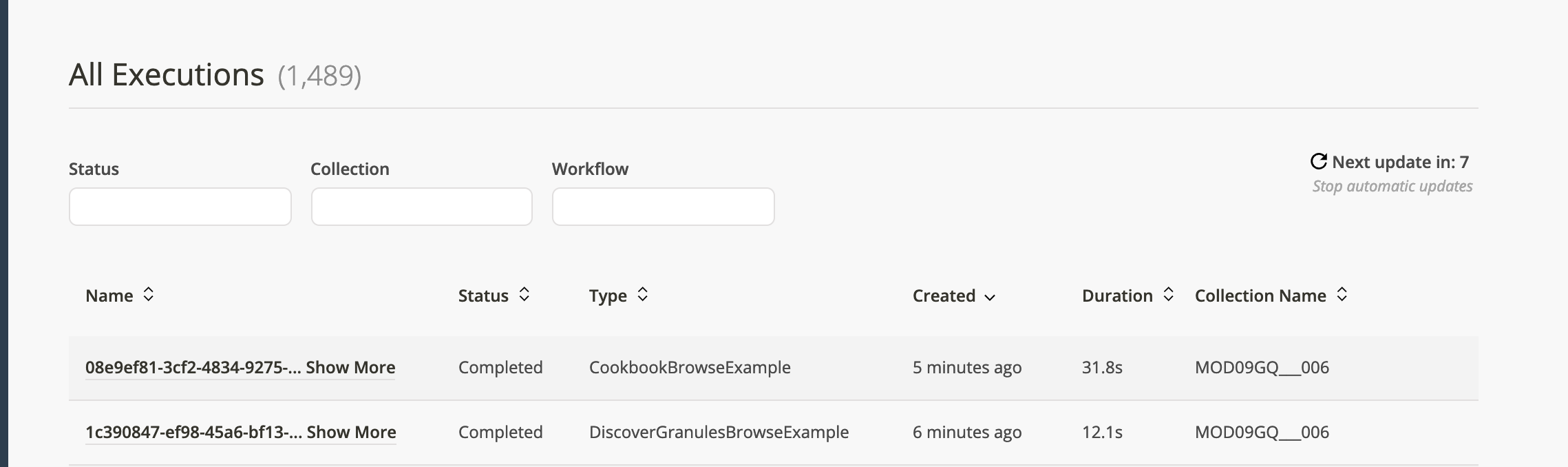
Tab over to executions and you should see the DiscoverGranulesBrowseExample workflow run, succeed, and then moments later the CookbookBrowseExample should run and succeed.
Results
You can verify your data has ingested by clicking the successful workflow entry:

Select "Show Output" on the next page
and you should see in the payload from the workflow something similar to:
"payload": {
"process": "modis",
"granules": [
{
"files": [
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"filepath": "MOD09GQ___006/2017/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"type": "data",
"bucket": "cumulus-test-sandbox-protected",
"filename": "s3://cumulus-test-sandbox-protected/MOD09GQ___006/2017/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"time": 1553027415000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{extractYear(cmrMetadata.Granule.Temporal.RangeDateTime.BeginningDateTime)}/{substring(file.name, 0, 3)}",
"duplicate_found": true,
"size": 1908635
},
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"filepath": "MOD09GQ___006/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"type": "metadata",
"bucket": "cumulus-test-sandbox-private",
"filename": "s3://cumulus-test-sandbox-private/MOD09GQ___006/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"time": 1553027412000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{substring(file.name, 0, 3)}",
"duplicate_found": true,
"size": 21708
},
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.jpg",
"filepath": "MOD09GQ___006/2017/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.jpg",
"type": "browse",
"bucket": "cumulus-test-sandbox-protected",
"filename": "s3://cumulus-test-sandbox-protected/MOD09GQ___006/2017/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.jpg",
"time": 1553027415000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{extractYear(cmrMetadata.Granule.Temporal.RangeDateTime.BeginningDateTime)}/{substring(file.name, 0, 3)}",
"duplicate_found": true,
"size": 1908635
},
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.cmr.xml",
"filepath": "MOD09GQ___006/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.cmr.xml",
"type": "metadata",
"bucket": "cumulus-test-sandbox-protected-2",
"filename": "s3://cumulus-test-sandbox-protected-2/MOD09GQ___006/MOD/MOD09GQ.A2016358.h13v04.006.2016360104606.cmr.xml",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{substring(file.name, 0, 3)}"
}
],
"cmrLink": "https://cmr.uat.earthdata.nasa.gov/search/granules.json?concept_id=G1222231611-CUMULUS",
"cmrConceptId": "G1222231611-CUMULUS",
"granuleId": "MOD09GQ.A2016358.h13v04.006.2016360104606",
"cmrMetadataFormat": "echo10",
"dataType": "MOD09GQ",
"version": "006",
"published": true
}
]
}
You can verify the granules exist within your cumulus instance (search using the Granules interface, check the S3 buckets, etc) and validate that the above CMR entry
Build Processing Lambda
This section discusses the construction of a custom processing lambda to replace the contrived example from this entry for a real dataset processing task.
To ingest your own data using this example, you will need to construct your own lambda to replace the source in ProcessingStep that will generate browse imagery and provide or update a CMR metadata export file.
You will then need to add the lambda to your Cumulus deployment as a aws_lambda_function Terraform resource.
The discussion below outlines requirements for this lambda.
Inputs
The incoming message to the task defined in the ProcessingStep as configured will have the following configuration values (accessible inside event.config courtesy of the message adapter):
Configuration
event.config.bucket -- the name of the bucket configured in
terraform.tfvarsas yourinternalbucket.event.config.collection -- The full collection object we will configure in the Configure Ingest section. You can view the expected collection schema in the docs here or in the source code on github. You need this as available input and output so you can update as needed.
event.config.additionalUrls, generateFakeBrowse and event.config.cmrMetadataFormat from the example can be ignored as they're configuration flags for the provided example script.
Payload
The 'payload' from the previous task is accessible via event.input. The expected payload output schema from SyncGranules can be viewed here.
In our example, the payload would look like the following. Note: The types are set per-file based on what we configured in our collection, and were initially added as part of the DiscoverGranules step in the DiscoverGranulesBrowseExample workflow.
"payload": {
"process": "modis",
"granules": [
{
"granuleId": "MOD09GQ.A2016358.h13v04.006.2016360104606",
"dataType": "MOD09GQ",
"version": "006",
"files": [
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"type": "data",
"bucket": "cumulus-test-sandbox-internal",
"filename": "s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"fileStagingDir": "file-staging/jk2/MOD09GQ___006",
"time": 1553027415000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{extractYear(cmrMetadata.Granule.Temporal.RangeDateTime.BeginningDateTime)}/{substring(file.name, 0, 3)}",
"size": 1908635
},
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"type": "metadata",
"bucket": "cumulus-test-sandbox-internal",
"filename": "s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"fileStagingDir": "file-staging/jk2/MOD09GQ___006",
"time": 1553027412000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{substring(file.name, 0, 3)}",
"size": 21708
}
]
}
]
}
Generating Browse Imagery
The provided example script used in the example goes through all granules and adds a 'fake' .jpg browse file to the same staging location as the data staged by prior ingest tasksf.
The processing lambda you construct will need to do the following:
- Create a browse image file based on the input data, and stage it to a location accessible to both this task and the
FilesToGranulesandMoveGranulestasks in a S3 bucket. - Add the browse file to the input granule files, making sure to set the granule file's type to
browse. - Update meta.input_granules with the updated granules list, as well as provide the files to be integrated by
FilesToGranulesas output from the task.
Generating/updating CMR metadata
If you do not already have a CMR file in the granules list, you will need to generate one for valid export. This example's processing script generates and adds it to the FilesToGranules file list via the payload but it can be present in the InputGranules from the DiscoverGranules task as well if you'd prefer to pre-generate it.
Both downstream tasks MoveGranules, UpdateGranulesCmrMetadataFileLinks, and PostToCmr expect a valid CMR file to be available if you want to export to CMR.
Expected Outputs for processing task/tasks
In the above example, the critical portion of the output to FilesToGranules is the payload and meta.input_granules.
In the example provided, the processing task is setup to return an object with the keys "files" and "granules". In the cumulus_message configuration, the outputs are mapped in the configuration to the payload, granules to meta.input_granules:
"task_config": {
"inputGranules": "{$.meta.input_granules}",
"granuleIdExtraction": "{$.meta.collection.granuleIdExtraction}"
}
Their expected values from the example above may be useful in constructing a processing task:
payload
The payload includes a full list of files to be 'moved' into the cumulus archive. The FilesToGranules task will take this list, merge it with the information from InputGranules, then pass that list to the MoveGranules task. The MoveGranules task will then move the files to their targets. The UpdateGranulesCmrMetadataFileLinks task will update the CMR metadata file if it exists with the updated granule locations and update the CMR file etags.
In the provided example, a payload being passed to the FilesToGranules task should be expected to look like:
"payload": [
"s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.jpg",
"s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.cmr.xml"
]
This list is the list of granules FilesToGranules will act upon to add/merge with the input_granules object.
The pathing is generated from sync-granules, but in principle the files can be staged wherever you like so long as the processing/MoveGranules task's roles have access and the filename matches the collection configuration.
input_granules
The FilesToGranules task utilizes the incoming payload to chose which files to move, but pulls all other metadata from meta.input_granules. As such, the output payload in the example would look like:
"input_granules": [
{
"granuleId": "MOD09GQ.A2016358.h13v04.006.2016360104606",
"dataType": "MOD09GQ",
"version": "006",
"files": [
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"type": "data",
"bucket": "cumulus-test-sandbox-internal",
"filename": "s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf",
"fileStagingDir": "file-staging/jk2/MOD09GQ___006",
"time": 1553027415000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{extractYear(cmrMetadata.Granule.Temporal.RangeDateTime.BeginningDateTime)}/{substring(file.name, 0, 3)}",
"duplicate_found": true,
"size": 1908635
},
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"type": "metadata",
"bucket": "cumulus-test-sandbox-internal",
"filename": "s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.hdf.met",
"fileStagingDir": "file-staging/jk2/MOD09GQ___006",
"time": 1553027412000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{substring(file.name, 0, 3)}",
"duplicate_found": true,
"size": 21708
},
{
"name": "MOD09GQ.A2016358.h13v04.006.2016360104606.jpg",
"type": "browse",
"bucket": "cumulus-test-sandbox-internal",
"filename": "s3://cumulus-test-sandbox-internal/file-staging/jk2/MOD09GQ___006/MOD09GQ.A2016358.h13v04.006.2016360104606.jpg",
"fileStagingDir": "file-staging/jk2/MOD09GQ___006",
"time": 1553027415000,
"path": "data",
"url_path": "{cmrMetadata.Granule.Collection.ShortName}___{cmrMetadata.Granule.Collection.VersionId}/{extractYear(cmrMetadata.Granule.Temporal.RangeDateTime.BeginningDateTime)}/{substring(file.name, 0, 3)}",
"duplicate_found": true,
}
]
}
],